Studying transcription in single cells
Interview with
Looking at large populations of cultured cells, it appears that when a stimulus is applied, genes are turned on and gene products are made continuously and in proportion to the intensity of the stimulus. But if you look at individual cells, a very different pattern emerges. National Cancer Institute researcher, Dan Larson spoke to Chris Smith...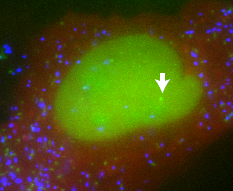
Dan - It turns out, and this is one of the punch lines of the paper, it seems like there are lots of ways for the nucleus to turn genes off. To turn the gene on requires lots of things to happen correctly. You need to recruit chromatin remodelers, you need to recruit what's called the basal transcription machinery. And we want to know how much of this one particular element was required for this gene to turn on. And the element that we looked at in this case was a steroid. We can quantitatively change the level of this steroid and we can look at the level of single RNA being transcribed to see precisely when transcription starts and how many RNAs are made.
Chris - So, the sort of premise here is that we know that when cells receive some kind of signal or they respond to some stimulus, the gene activity can be changed. Genes can be turned on.
Dan - Correct.
Chris - But what we're not so clear on is how the cells respond in terms of how much RNA, how much gene transcription goes on, in response to any given stimulus.
Dan - Yes. One of the ideas to emerge in recent years is that most RNA is not made in continuous levels. When you look in a population, you see this nice smooth increase in the amount of RNA may have the function of the amount of the upstream signal present. But when you look at a single cell level, RNA is transcribed in these short bursts of transcription. Punctuated by very long pauses where no RNA is made.
Chris - If we were to zoom in on the genome at the time that's happening, does that reflect assembly of transcription machinery or something which is why everything is there, all the ducks are in a row, it gets transcribed that then things stop while it resets itself or is there some other fundamental reason why you should get that very staccato, lots, nothing for a while, lots sort of phenomenon?
Dan - That's a good question. I would say in many cases, we don't know what's happening at the molecular level. What we know is that for example for this genome we studied in this paper, it transcribes for about 30 minutes and then it goes off for about 30 minutes even though the upstream signal isn't changing at all. Now, the timescales of this are suggestive possibly of chromatin remodelling events which may control some of the timing of this transcriptionional bursting or maybe due to the stability as you say of some of these factors at a promoter.
Chris - So, how did you try to get forward on this and get a handle on what was going on?
Dan - So, what we did is, we used a system that allowed us to visualise RNA in living cells. We make use of a system that mother nature has already used - the packaging of RNA into a bacteriophage. There's a bacteriophage capsid protein which binds a stem-loop and then that RNA containing a stem-loop is packaged into a particle. We have adapted that system and we use the same stem-loops and we encode them into our gene of interest. At the same time, we have capsid protein that's expressed in the cell and it's labelled a fluorescent protein. So once these stem-loops get made, the capsid protein binds and now, we can see this RNA in the microscope.
Chris - Wow! So, you can just count the particles that get made and that tells you how many RNA transcripts were coming off of that gene and see one time. What a phenomenal way of doing it. So, what have you learned?
Dan - So, what we've learned from that is that even when this gene is transcribing at saturation, there are long periods where no RNA has been made, again, due to this bursting phenomenon. When we increase the level of the steroid, the nuclear is set to respond to that steroid and a gene which responds to that steroid changes the frequency of this bursting but doesn't change the length of the burst and doesn't change the amplitude. So, what you can think of this is a toggle switch that's going on and off. Steroids control the frequency of this toggle switch.
Chris - So, what do you think are the implications of this? How does this inform or change what we thought before? Why is this important - the whole phenomenon?
Dan - I think one of the things that really comes out of this study is that we took a very well-understood physiological response. The response of a steroid responder gene to steroids. Biochemistry of these systems has been studied for 60 years and the dose response is one of the most well-characterised dose responses in pharmacology. It's a billion dollar pharmaceutical industry, understanding how steroids affect gene regulation. All of these studies are predicated on this idea that cells respond uniformly. That these dose response is due to the affinity of the receptor and the ligand. What we have shown is that when you look at a single gene level, you see something completely different. So, what we see is this complex frequency modulation which would never have been guessed based on ensemble studies and which suggests that this basic dose response mechanism may be way more complicated than we originally thought. It also raises the possibility that changing the dynamics of a ligand - in this case, the steroid - could lead to newer improved ways to use steroids as therapeutics.
- Previous Introducing the Brain Panel
- Next Cell death at a distance









Comments
Add a comment