Drug Discovery: On this week's Naked Scientists, drug development goes under the microscope as we explore two new ways to find the treatments of tomorrow. We find out why size is important when it comes to chemicals that can kill superbugs, and how soil bacteria hold the chemical clues to the next generation of antibiotics. Plus, how sheets of carbon can be used to reveal single atoms of hydrogen beneath the microscope, how scientists have homed in on the part of the brain linked to obsessive compulsive disorder, and why electronically tagging jellyfish can tell us a sea turtle's secrets. Plus, in Kitchen Science, we make strange and unexpected shapes with bubbles!
In this episode

- Could drugs evolve like diseases?
Could drugs evolve like diseases?
We put this question to Dr Melanie McCullagh:
Melanie - That's a very interesting question. I'm not sure that you would actually want a vaccine to evolve too much just in case once you set it loose in the wild you would risk some kind of a vaccine epidemic. I think what you can definitely do is apply selection pressure to the bacteria to change the types of molecules that they produce. That's a similar kind of concept. Chris - I would think that probably this is going to be big business in the future. We're only just beginning to understand how molecules actually look. By turning this round in this way we can now start with what we want to end up with and then go back to the gene and tweak that. That's quite a novel thing to be doing. Probably going to be a major way of doing this in the future. Melanie - There are some real advances in technology at the moment that I think will enable us to expand greatly what we can do with genetically engineering the bacteria that produce these polyketides. Biotica's a very small company but I think there's a really big area for us to build into with increasingly novel technologies as things move on.

Smallest atoms seen for the first time
A team of US scientists have achieved a genuine quantum leap in imaging - with the development of a technique that's enabled them to see, for the first time, the smallest atoms, including hydrogen. 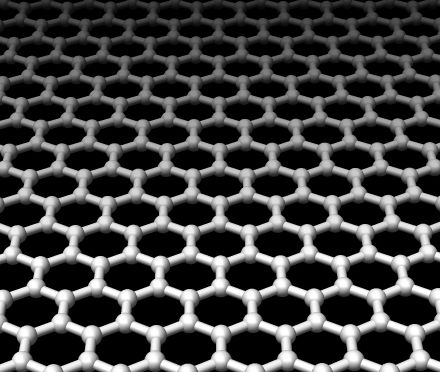
Writing in this week's Nature, UC Berkeley researcher Jannik Meyer and his colleagues placed a sheet of graphene, which is a single layer of carbon atoms arranged in a honeycomb configuration beneath an electron microscope. In the electron beam they were able to pick out tiny dots, like stars in a cloudless sky, which were individual atoms of various types that had landed and stuck to the graphene sheet.
Incredibly, alongside the carbon atoms that were relatively easy to spot, were some fainter smaller dots which fitted the bill both electrically and in terms of their appearance to be hydrogen atoms. The team were also able to image whole molecules including hydrocarbon chains, which as one might predict formed lines across the image.
The amazing results are thanks to the choice of graphene as the backdrop for the imaging. Being one atom thick and highly regular in its configuration it's very easy to subtract electronically from the signal to leave just the image of the atoms. Graphene is also very sticky, so it holds onto the atoms and molecules that land on it very tightly, and it is also very robust so it can withstand the intense beam of electrons hitting it.
At the very least this technique promises to open up new vistas in imaging, including the ability to study the shapes and arrangements of atoms in complex molecules.
Reference: Nature 454, 283-284 (17 July 2008) | doi:10.1038/454283a; Published online 16 July 2008

The details in the devil - how a tumour is driving a sexual shift in Tasmania
Researchers in Australia have shown that the Tasmanian Devil, beleaguered in recent years by a transmissible fatal facial cancer, has responded to the disease threat by a dramatic change in its reproductive habits. 
Writing in this weeks PNAS, University of Tasmania scientist Menna Jones and her colleagues looked at devil population data from five locations around Tasmania. Prior to the appearance of devil facial tumour disease, the data shows, the animals lived an average of 5-6 years, began breeding from the age of two years and raised several litters of offspring over their lifetimes.
Now the animals, numbers of which have fallen by nearly 90% since the disease first appeared in 1996, are breeding once, at less than a year old, and may not even survive long enough to rear that litter.
The reason for this precocious sexual maturity isn't known, but scientists suspect that a combination of factors including less competition from other devils, reduced population density and more plentiful food may be responsible. What it means to the devils is even more unclear, although it could prove to be their salvation if it leads to the emergence of animals that are resistant to the disease, which is thought to behave like an infectious organ transplant spread when the animals fight and bite one another.
However, there are very few devils left and their genetic diversity across Tasmania is generally very low, which means that will be difficult for natural selection to pick out animals that are resistant to the disease and for these animals to replace the population. Consequently the team are not optimistic, sombrely suggesting that the animals may be extinct in the wild within 25 years.
"This novel disease could have catastrophic consequences for the Tasmanian devil," they said.
Reference: Published online before print July 14, 2008, doi: 10.1073/pnas.0711236105
Obsessive research highlights cause of OCD
Scientists in Cambridge have pinpointed changes in brain activity underlying obsessive compulsive disorder. The discovery could lead to more accurate and earlier diagnosis of the disabling condition, which affects between 1-3% of the population, runs in families, and is associated with frequent intrusive thoughts (obsessions), which provoke sufferers to perform recurrent rituals (compulsions) such as hand washing or turning lights on and off. These symptoms can be very distressing and can take up hours of a patient's time. They're often also diagnosed late, which might also make them harder to treat when the sufferer eventually seeks help. 
But that could be about to change; writing in this week's Science, Sam Chamberlain and his colleagues recruited 14 people suffering from OCD, 12 of their immediate relatives, and a control group of 14 unaffected individuals. In a brain scanner the volunteers were shown two pictures side by side of a house and a face. They were then asked to press one of two buttons to indicate which of the two images was the "correct" answer, which they had to work out initially by trial and error. After the correct "target" had been identified six times in a row it changed, so that the subject had to learn again.
The researchers then compared the patterns of brain activity between the subjects and found that amongst the OCD sufferers and their relatives a brain region called the lateral orbitofrontal cortex was consistently less active. This region is concerned with reacting to change, which explains the ritualistic behaviour shown by OCD sufferers.
"Impaired function in brain areas controlling flexible behaviour predisposes people to developing the compulsive rigid symptoms that are characteristic of OCD," says Chamberlain. "This study shows that these brain changes run in families and represent a candidate vulnerability factor. The current diagnosis of OCD is subjective and improved understanding of the underlying causes of OCD could lead to more accurate diagnosis and improved clinical treatments."
Reference: Science 18 July 2008: Vol. 321. no. 5887, p 421 - 422 DOI:10.1126/science.1154433

Where does the milk in a coconut come from?
There's an issue here because coconut milk is actually slightly different to coconut water. It's coconut water that you find inside a coconut. That's not water that's got in from the outside, it's created by the coconut itself. It's actually what we call the endosperm of the coconut plant. That's the stuff that gives the nutrition for the developing seed, a bit like the flesh of a banana. As the fruit ripens, the coconut water gets converted into the solid fruit that you find in there. If you have an unripe coconut, the water will be quite sweet and very nice. The riper it gets the less pleasant it gets to drink.
Are there any hereditary psychiatric conditions?
Yes. Schizophrenia - we know that the risk rises very much so if you have a first degree or near-relative with the condition. Also bipolar disorder. They're two very common conditions. They affect between one and two percent of the population.
Scientists are now beginning to apply the power of molecular biology to try to find out what the genes are that are linked to these conditions. They're using something called single nucleotide polymorphism analysis or SNP analysis. What this involves is a big group of people who have the disorder, a group of people who don't have the disorder and then you look across their DNA for these little molecular markers called SNPs to see if they crop up more often in certain places of the genome in people who have a condition than people who don't. That gives you a kind of flag sign or a way-marker to say this bit of the DNA might be important in this condition. Then you can focus your attention much more closely in that area. The same analysis has proved that there are various other genes that, in addition to the ones we knew about, are linked to things like genetics for breast cancer and diabetes in the last twelve months. I think we'll probably see quite a lot of fruit being borne out by that procedure quite soon.

Will an iPod weigh more when it's full of music?
It will, Neil, if you fill it with heavy metal (!)
But really, the answer's no.
The way in which an iPod works depends on which one you've got. If you've got the one which uses solid state memory then all it's doing is storing binary data. It's using a memory chip which is storing information as digital information.
If you've got the older, bigger iPods that have hard-discs in them, this is magnetic binary data. But in either case it's storing a "1" or a "0" by having something - such as a piece of magnetic material - pointing one way or the other.
So it doesn't actually matter whether it's actually got anything stored on it or not, because storing nothing still weighs the same as storing something. It's not like a cupboard that you're putting tins into.
On our forum, Madidus Scientia put it very well saying, "it's a bit like having a handful of coins and they're all either heads or tails." That's like the "0" or "1" in digital binary. They weigh the same whether they're all showing heads or they're all showing tails.
So there's no reason to think there should be a difference in the actual weight.
However, when you charge the battery, you are adding energy and there is a relationship between energy and mass...
As Einstein said E=mc2, so when you boil your kettle or - and this is the best excuse for not doing P.E. at school - when you run, in both cases (the hot kettle or you running) you have more energy.
When you're running faster you have more kinetic energy. Because E=mc2 - that's E, energy, equals m, mass, times the speed of light, c, squared, and since the speed of light, c, doesn't change, if your E, energy, goes up, your mass must go up. So a hot kettle will weigh more, and when you run in P.E. you will gain something like 10-14g. This is not a prodigious weight-gain but it is nonetheless weight gain due to taking exercise!
Similarly, in your iPod, when you charge it up you're putting energy into the battery and it will weigh a tiny amount more. In fact, one statistic we did hear is that a thumbprint applied to the front of the iPod in the form of, say, the grease on your thumb will weigh thousands of times more than the increase in the weight of the battery due to charging it...

27:19 - The Search for New Drugs
The Search for New Drugs
with Dr Harren Jhoti, Astex Therapeutics
Chris - Joining us in the studio this week is Dr Harren Jhoti who is the founder of the drug discovery Astex Therapeutics. He set this up in 1999. He started off as the chief scientific officer and he's now the chief executive officer. Harren, thank you for joining us. What's the basic way in which, before you came along with your techniques, we were trying to invent new drugs?
Harren - The conventional approach to discover new drugs really starts with initially finding a target which may be associated with a particular disease. That target, quite often, is a protein which malfunctions in one way or another. Sometimes it's hyperactivated the protein's function. What you then do is you take that protein and then you screen it against a collection of compounds. Basically these are libraries of chemical entities. Quite often these libraries are really rather huge in the sense there could be hundreds of thousands of different compounds. The aim there is to identify some compounds which actually bind or interact with that target protein. Those are called leads in the drug discovery terminology. After that what happens is you try to optimise those leads by doing some iterative chemistry, i.e. you try to improve the interaction of those compounds against the target protein. You try to impart in those compounds the drug-like properties one needs for that compound then to become a drug.
Chris - In the real world how long would it take between you coming up with a structure for something in the body you want to target. Say you find a gene which makes something linked to, say, high blood pressure and you want to make a drug to lower blood pressure by blocking that gene. How long would it take if you invented a molecule between the invention of that molecule, the conception in the laboratory, and it actually going into a patient? What's the average time?
Harren - You know, this process and procedure is a really very long time. It takes many years and it ranges between five to ten years often to go from the concept to a drug which may get onto the market place and to treat patients. That's why it requires a huge amount of investment in this whole process.
Chris - What's the average price tag for a drug coming out today?
Harren - The kind of monies it requires to develop a drug is in the order of $800 million.
Chris - Wow.
Harren - That number also does include a lot of the programmes which actually fail to generate the compound.
Chris - So that's why pharmaceutical companies when they make a drug have to charge a lot for it? Certainly to the first world, in order to make their money back to bankroll the development of other drugs that are not going to succeed?
Harren - That's exactly right. The actual failure rate is really quite eye-watering, in fact. Depending on where you look at the process, what the metrics are. They range in the order of one in 100 or one in 1000 compounds actually get to the market place.
Chris - Why do most not make it?
Harren - That's a big question and a lot of people are trying to understand how we can improve this productivity. One thing which is quite clear from looking at the types of chemistries which have been generated using this more conventional approach is that perhaps these molecules have become too molecularly complex. Perhaps they're a little bit too large. What happens then in the body is that some of these compounds are actually metabolised in a negative way which generates toxicities.
Chris - So you're saying the actual drug that the company might make ends up being far too big and ends up turning into other things in the body which can have harmful effects. That's why the compound fails?
Harren - There is clear evidence which suggests that the attrition or the failure rate of compounds which are being developed in the clinical setting is correlated with the size of the molecule.
Chris - Do you think it's also correlated with the size of the legal bills that companies like Merck have been facing? When they come up with a drug and then get sued for a billion dollars a few years later because of consequences which came to light later this is probably a massive disincentive, isn't it?
Harren - There is no question these are broader societal pressures on the industry. I think in a very general way society has perhaps become a little bit overly risk-averse. There's no question every drug will be toxic to some degree, depending on the dose. The whole question here, or the challenge, is to give appropriate amounts of drug to deal with the disease and not generate toxicity.
Chris - What's the approach you've been taking at Astex to do it differently and surmount some of these difficulties?
Harren - The key issue that we've been trying to target is to try to keep the eventual drug candidate, the drug molecule, slightly smaller - slightly less complex. What we do is, rather than screen these compounds which are quite often between 300-500 Daltons in size we actually screen much smaller chemistry: between 1-200 Daltons. [1 Dalton is approximately the mass of a hydrogen atom] These are what we call fragments. Then what we're able to do is add chemically to those fragments to improve the interaction of those fragments to the target protein.
 Chris - Still sounds like an enormous amount of work, though. How are you able to do this and do it more efficiently and more cost-effectively than other companies? If it was that easy they'd be doing it.
Chris - Still sounds like an enormous amount of work, though. How are you able to do this and do it more efficiently and more cost-effectively than other companies? If it was that easy they'd be doing it.
Harren - There's no question it is difficult. What we've been doing is focussing on how you would optimise the technology to be able to firstly detect these very small fragments which is a technical challenge in itself and then to optimise. What it really boils down to is visualisation of how these fragments bind. We've developed biophysical techniques: x-ray crystallography, NMR and integrated the suite into an approach which we call Pyramid to allow us to do this fragment-based drug discovery.
Chris - Talk us through the Pyramid bit-by-bit, exactly how you would go about your technique for making a new agent.
Harren - Once we've identified the target protein we then produce the three-dimensional structure in that protein by doing x-ray crystallography. For that you have to do a lot of protein expression and biochemistry.
Chris - So that gives you the shape and what the molecule actually looks like. How does that help you?
Harren - That helps in two ways. Firstly, it tells us exactly where a fragment has been bound to that target and that allows us to rationally optimise that fragment to fit better into that target protein.
Chris - So that gives you the business-end of the drug so you know which bit is doing the important job?
Harren - That's right. Then what we could do really is what we're doing is hand-crafting these molecules to very uniquely directly fit into the pockets of the protein. The other key advantage of his approach is that these fragments are very low affinity. They bind with very low interactions and it turns out that biophysical techniques such as x-ray crystallography and NMR are really much better ways of detecting the original binding of the fragments.
Chris - Then where do you go?
Harren - What we then do is do several cycles of optimisation. We add extra functional groups onto these fragments and grow these fragments out, very much like a seed. Like planting a seed and growing a seed. Once we actually have molecules which have the appropriate profile in terms of the properties one requires from a drug transit these molecules are then tested in human beings. Of course before you do that you have to test them in preclinical models.
Chris - Once you're arrived at this molecule you want to make how do you actually get it so you can make it efficiently? If you end up with very complicated chemistry and needing to make these very complicated but small molecules, how do you make them?
Harren - It turns out actually the fragments are very simple, much more simple, than the larger complex molecules which conventional screening or conventional drug discovery actually use. The chemistry's simpler and less chemically challenging than conventional drug study.
Chris - Have you got drugs that are actually out there in the clinic or which are just experimental at this stage?
Harren - We've put three compounds now into clinical trials. Our particular focus today is developing the experimental cancer therapies. We have three compounds being tested in cancer patients: both in the US and in the UK.

36:38 - Discovering Drugs from Bugs
Discovering Drugs from Bugs
with Melanie McCullagh, Biotica Technology
Chris - Welcome Melanie, thank you for joining us. What are you actually up to?
Melanie - Yes, Biotica has a technology which I would say is quite the other end of the spectrum from Astex. Polyketides that are a type of compound produced naturally in soil bacteria. A lot of them are very naturally pharmaceutically active as well.
Chris - What are they?
Melanie - They're fairly large molecules. They're around 500
Daltons and they are made up of a series of carboxylic acids which are joined together in the bacteria by an enzyme called polyketide synthase.
Chris - What do they do?
 Melanie - Well, we're not completely sure what they do necessarily within the bacteria. We don't know why they produce them per se but when we give them to humans they interact with a really wide range of targets. Some of the really famous examples, for example rapamycin, is an immunosuppressant. It's used in transplant patients to prevent them rejecting the organ that's been given. There are also analogues of rapamycin which are used in cancer, for example. It's been used and tested in other indications as well. They're more broadly applicable than that. There are antibacterial polyketides such as erythromycin. Some of the early anti-cholesterol statins were also polyketides and they're also used in cancer. Some really novel cancer compounds which have been recently approved are also polyketide compounds.
Melanie - Well, we're not completely sure what they do necessarily within the bacteria. We don't know why they produce them per se but when we give them to humans they interact with a really wide range of targets. Some of the really famous examples, for example rapamycin, is an immunosuppressant. It's used in transplant patients to prevent them rejecting the organ that's been given. There are also analogues of rapamycin which are used in cancer, for example. It's been used and tested in other indications as well. They're more broadly applicable than that. There are antibacterial polyketides such as erythromycin. Some of the early anti-cholesterol statins were also polyketides and they're also used in cancer. Some really novel cancer compounds which have been recently approved are also polyketide compounds.
Chris - What you're saying is that bacteria have a pretty broad medicine chest at their disposal. All we have to do is to work out how to get it out.
Melanie - Yes, that's right.
Chris - What are you doing to do that?
Melanie - Biotica specialises in genetically engineering the bacteria so that we can change the properties of the polyketides that are naturally produced. One really good example is there's a polyketide called FK506 which is an immunosuppressant. It's used in transplant patients. What we're doing is working on that molecule to change its pharmaceutical properties, make it appropriate for delivering for example by inhalation. We can then take advantage of the fact that it down-regulates inflammation, down-regulates the immune system and give it to asthma patients. The interesting thing about FK506 is that when it's given systemically it's quite toxic. It has very variable metabolism. What we can do it we can change the metabolism of the drug to make it more appropriate to give in a bigger indication like asthma, really reduce the toxicity of that molecule. 
Chris - How do you how to tweak the DNA of the bacteria so that they make a different molecule which is more human-friendly?
Melanie - The polyketide synthase is a very interesting enzyme. The genes directly translate to different molecules within the polyketide synthase. Each of those will produce carboxylic acid and may change the acid as it's added onto the polyketide. By looking at the genome with the bacteria we can actually identify how to change the individual modules within the polyketide synthase. For example, we can swap from one organism to another to recruit different acids into a polyketide molecule. We can tell by looking at the structure and activity relationships of the molecules that we know we can tell what changes we have to make to make changes in the activity of the molecule within the human.
Chris - Is anyone else doing this or is this your unique selling point?
Melanie - Biotica has patents on this technology. We're the only company who is able to do certain parts of the technology. There are other companies that are doing similar things, perhaps with different types of molecules. There was another company who had some other kind of overlapping technology in the US which has recently been bought by one of the very big pharmaceutical companies.
Chris - So the bottom line here is that, rather than taking the molecule and fiddling with it chemically, you can actually take a molecule that the bacteria are already making, fiddle with the bacteria and make them make it better?
Melanie - We can get the bacteria to do the work for us. The bacteria also produce the molecules commercially as well. We can scale-up, ferment lots of bacteria and get pharmaceuticals out really efficiently.
Chris - Because bacteria don't charge a salary and they don't get bogged in health and safety?
Mario - Is it possible to create an intelligent vaccine or a drug that could evolve with the ever changing strains of the disease?
Melanie - That's a very interesting question. I'm not sure that you would actually want a vaccine to evolve too much just in case once you set it loose in the wild you would risk some kind of a vaccine epidemic. I think what you can definitely do is apply selection pressure to the bacteria to change the types of molecules that they produce. That's a similar kind of concept.
Chris - I would think that probably this is going to be big business in the future. We're only just beginning to understand how molecules actually look. By turning this round in this way we can now start with what we want to end up with and then go back to the gene and tweak that. That's quite a novel thing to be doing. Probably going to be a major way of doing this in the future.
Melanie - There are some real advances in technology at the moment that I think will enable us to expand greatly what we can do with genetically engineering the bacteria that produce these polyketides. Biotica's a very small company but I think there's a really big area for us to build into with increasingly novel technologies as things move on.

42:38 - Are any organisms immortal?
Are any organisms immortal?
We spoke to Dr John Nudds, Senior Lecturer in Palaeontology at the University of Manchester to find the answer...
One of the longest living vertebrate animals, and many listeners will be aware of this, is the giant land tortoise. There's a nice story about Captain Cook, the explorer, presenting one of these animals to the queen of Tonga in 1788. This animal eventually died in 1966, 188 years later. The animal was probably mature by the time he collected it. If we turn our attention to the plant kingdom we can multiply these figures by a factor of ten. There's a well-known example of the bristlecone pine trees which grow in the rocky mountains of North America. These are well-known to live for over 4000 years. I think the record's about 4600 years.
Again these figures have recently been doubled by research in Sweden. Scientists here came across a Norway spruce whose root system had been growing for 9550 years!
If we now move onto some of the simpler life forms then the numbers do start to get really big. In 1995 a sample of bacteria was found in a stomach of a bee which was encased in amber which was dated at between 25 and 40 million years old. These bacteria were found in a state of suspended animation. They had to be reanimated in the laboratory. In scientific terms they were in what we would call a crypto-biotic state. It means the cells remained alive but none of the life processes were being carried out. They didn't feed or reproduce so whether you consider this as immortality or not is open to question. To answer the question, the sad fact is that all cells do decompose with time. All cells age and all cells eventually die so sadly, as yet no life form has evolved that is immortal.

What is a Dalton?
We put this question to Dr Harren Jhoti:
Harren - Basically a Dalton is just a measure of weight, it's a measure of mass. It's a very, very small measure of mass: something like micrograms or grams people refer to but at a kind of microscopic level, if you like. Daltons are basically very single units of small mass.
Chris - So when you're down at the molecular level you can't use grams or even thousandths of grams? You need to use much smaller molecular units?
Harren - That's exactly right. That's the most commonly-known unit which people refer to when looking at small molecules.
Are there different body types when responding to drugs?
We put this question to Dr Harren Jhoti...
Harren - Yes, I think this is a very interesting question. It really touches on what's happening in a broader sense in the pharmaceutical industry. People are really talking about personalised medicine.
This issue that body types or different types of people, often due to their different genotypes or genetic make-up - it's going to be very interesting to see how drugs actually are metabolised by different types of people and whether those drugs show different levels of efficacy in different types of people.
There's a huge push in the industry now to see whether we can't try to predict up front by reading the genotype of a particular person and whether that person is going to a) respond to the particular treatment and b) whether they're going to have a higher chance of having detrimental side-effects due to some toxicities.
There's a big push in this whole personalised medicine. It's based on body types and genome sequencing. It's all part of the same issue.
Chris - It's very interesting, isn't it. If you go into a shoe shop you expect to get a pair of shoes that's the right size for you. But you go into a chemist and you're given a drug which millions of other people take and it's expected to fit your molecules in your body just perfectly, even though you might be totally different from the next person.
Harren - Yeah. That's a reality today because we simply haven't had the technologies to be able to segregate people into these different groups. Genome sequencing is a very recent development and many of these drugs were actually discovered and developed ten or fifteen years ago. I think going forward molecularly-targeted agents which try to target subgroups of patients is going to be very much the focus.
How do you train antibodies to attack skin cancer?
We put this question to Dr Melanie McCullagh:
Melanie - Monoclonal antibodies are a very interesting new way of treating cancer and one of the really clever things about them is that they target specific receptors on the surface of the cancer. What we know about antibodies is that their structure is like a lock and key. They fit very closely onto the receptor and are very specific to where they take the activity of the antibody. In melanoma, which is a particularly serious form of cancer with some really quite poor outcomes, there's some really great work going on at the moment looking at making the antibodies target those receptors much more specifically and much more clearly to the cancer itself. Chris - Do you see this as being a massive growth area now? Are we going to see people that are better at doing this in future?
Melanie - I think it's really important and there's a lot of companies working on how to develop new forms of antibodies with a greater specificity for cancer but also with different types of activities. Even ones that can carry other cancer therapeutics directly to the site of the tumour, which is very interesting.
Chris - It's very important too since one person in every three will ultimately in their lifetime die of cancer.










Comments
Add a comment