It's thought that around one in a hundred children in the UK is somewhere on the autistic spectrum - a complex range of disorders that can be challenging to understand and live with. But recent advances in genetics are bringing hope for new therapies that might help. Plus, we look at the genes underlying Specific Language Impairment, find out why cancer has the X factor, and meet a hopeless-sounding gene of the month.
In this episode

01:07 - Dr Steve Scherer - Autism genetics
Dr Steve Scherer - Autism genetics
with Professor Steve Scherer, Hospital for Sick Children, Toronto
We're all becoming more aware of autism and related disorders such as Asperger's syndrome, and more than 100,000 children in the UK are currently thought to be living with an autistic spectrum diagnosis. The causes of autism are not fully understood - although any link with childhood vaccines has been ruled out by robust scientific studies - but it's clear that genes play a role.
To find out more about the complex genetic landscape of the autistic spectrum, I spoke to one of the world's leading experts in the field, Dr Steve Scherer at the Hospital for Sick Children in Toronto. I started by asking him to explain what autism actually is.
Steve - Autism is a childhood developmental disorder that actually lasts a lifetime, that involves deficits in learning, repetitive stereotypic behaviours, anxieties, and language communication deficits. So, these are grouped into a so-called triad of symptoms that lead to an autism spectrum disorder diagnosis.
Kat - So, what do we know so far about what causes autism?
Steve - So, there's been many studies over 40 or 50 years in fact, probably the most interesting data came in studying identical twins that have the same genomes. And if one sibling twin was on the autism spectrum, in some of the studies published, over 90% accordance rate or chance that the second twin would also be on the autism spectrum. And then in the last say, 8 years now, but more in the last few years, the ability to scan DNA at a very high resolution using what we call 'genome technologies' and more recently, by doing genome sequencing itself. So, you're decoding all of the chemical bases of information in the genomes of individuals and we've been able to find some of these genes that are in fact causative for autism.
Kat - With a disease that's as complex as autism and there is a wide spectrum so people can have different levels of it, if you like, it seems obvious to me that it's not just one gene. What do we know about the genes for autism so far and how they interact?
Steve - So, what's really come out in the last few years is the fact that we now know for sure there's dozens and dozens, and dozens, and we estimate perhaps a few hundred genes that can be involved in this very complex clinical disorder that we were calling autism spectrum disorder. If you look at the clinical expression of the disorder in identical twins that have the same genome, in some cases, you could have one child who's very mildly affected with say, Asperger's syndrome and another identical twin who's severely autistic. So, this gives kind of information that there is this underlying clinical complexity and we also now know the genetic complexity. There are certainly at least a dozen or so bona fide genes that have been identified that if you have only one copy of that particular gene instead of the typical two, there's a very high probability that you'll be on the autism spectrum.
Kat - And what sort of things, certainly of the genes that we know about already, what sort of things do they do in the body?
Steve - So, there were a lot of hypotheses before about what genes might be involved and they were completely wrong to be honest. But in retrospect, the genes that have been identified, it makes sense. It's just that people were kind of chasing the wrong ideas. So, the genes that have been identified are encoding proteins that are involved in how the brain nerve cells or the neurons develop and communicate with each other. They're so-called synaptic genes. They work at the synapse or the area where the neurons come together to exchange information. And what's very interesting is, most of the autism, what we call 'susceptibility genes' found now, encode molecules that are working together in what we call a genetic pathway or a biological network at the synapse in the brain. And that becomes really important because if you have a very genetically heterogeneous disorder like we know now in autism, we'd like to try to use that genetic information to help us guide development of therapeutics - medicines, drugs. But if each one of these variants only accounts for 0.5% or less, it's pretty hard to develop hundreds of different drugs. But you might imagine, if they're all working in the same pathway, different components of pathways, you might be able to group together or stratify patients into certain groupings that may - say, drug A may have an impact on them, or drug B may impact another group of patients. And that's kind of the underlying idea going forward.
Kat - Because that's a very exciting concept and it's certainly for families that are affected by this condition because there could actually be drugs that will be effective in the future.
Steve - Yes, so right now, there is no effective drug in any individuals as far as we know for the core deficits that are involved in autism. Some of the medically associated observations like anxiety, we see in a very high percentage ADHD for example, you can take some drugs. In some cases they work, in some cases, they don't. Some cases, they have adverse effects, but not for the core symptoms for autism. So, that's precisely the approach. What typically happens is when a gene is identified, we can then go and generate the equivalent of genetic change or mutation in an animal model - we usually use mice for this - check to see if the mice have characteristics like autism. We'd look to see how they groom themselves, how they interact with their littermates, things like this.
And then you can test drugs on these mice and in many cases, some drugs have been developed for different things that are known to work on these proteins that we know now are involved - genes or proteins involved in autism. So, we can test those or we can develop new ones. There's been quite a bit of progress. There's at least 6 or so different drugs that have been developed, in particular for other medical genetic disorders like Fragile X syndrome and Rett syndrome, of which a high proportion of patients actually have autistic-like characteristics too.
Those genes were identified 20 years ago and they've gone through this whole route of looking at animal models and generate drugs. It turns out, those drugs can also be tested in autism populations. If you look at that group of the population, it has mutations in genes that are involved in the Fragile X and the Rett syndrome pathways, and that's exactly what's happening. And then for some of the newer genes that are involved in different types of synaptic proteins, there are some other off the shelf drugs that have been developed in other studies, for example in ADHD, that can now be tested in the population of autistic individuals that have their alterations in that genetic pathway. So, that's exactly what's happening.
Kat - It sounds like the next couple of years are going to be an incredibly exciting time in this area.
Steve - It is. It's kind of opened up the black box now and we've shed light on what the real problem was and the biological challenges so we can do experiments that are informed and design things in a right way. So, we're seeing incredible progress, but the key will be to turn that information to benefit individuals involved and the families so that they can have a better life.
That was Dr Steve Scherer at the Hospital for Sick Children, known as Sick Kids, in Toronto, Canada.
08:22 - RNA goes loopy
RNA goes loopy
with Nell Barrie
Nell - Well, I like this first one because it's got the headline, "RNA goes loopy" which sounds like something you get in a tabloid, but they probably wouldn't be covering this type of thing.
Kat - "Goes crazy kills everyone!". What's RNA up to now?
Nell - So, as you might have guessed from "RNA goes Loopy", this is about RNA that's circular. So, normal idea, sort of GSE students looking at "the messenger" of the cell - RNA is like a little string. But in this case, we're talking about circular RNA which we didn't really know was a possibility or existing until quite recently.
Kat - So, why haven't we found these circular RNAs before, because it seems completely contradictory to everything we know about RNA so far?
Nell - Well, yeah. I found this interesting because actually, it seems like the fact that we were finding all these linear RNAs, it was actually an artefact of the way that researchers were doing this type of study because the way that you find them is by looking for characteristic tails or these little molecules. Obviously, if you've got a circle, it doesn't have a tail so you're not going to find it.
Kat - You don't find what you're not looking for?
Nell - Exactly, yeah.
Kat - So, this is really intriguing because what are these up to? I mean, last, last month, we talked about the new DNA, this quadruplex DNA. Now, we're talking about circular RNA. Is it actually important?
Nell - Yeah, I mean, the quadruplex genetic, it's so last week, it's all about loopy RNA this week? Really, what we're looking at is what these little funny structures could be doing inside the cell and as any good biologist will know, it's all about structure and function. So, the fact that they're circular could mean that they're interacting in different ways with things inside the cells and that's really what researchers are looking at at the moment.
Kat - And it is important to point out that these are naturally occurring, so it's likely that there's probably a whole bunch more in our cells that we don't know about. It's two absolutely fascinating papers that came out in Nature this week. And then I guess there's a whole new field of RNA research opened up here.
Nell - Yeah, absolutely and what this shows is that the researchers have actually gone back and looked at previously existing databases and found new examples of circular RNA that they didn't know was there before. And it's looking like it could be doing interesting things like mopping up other RNAs that are in the cell, perhaps interfering with some types of processes, looking at things like binding with proteins from viruses, all that kind of stuff. So there's a lot more research to be done to find out exactly how these things work.
10:49 - Cancer gets the X factor
Cancer gets the X factor
with Nell Barrie
Kat - And another story that we saw this week was published in the journal Cell from Jeannie Lee and her team in Boston, finding potentially a use for another RNA called Xist in blood cancers. Now, you've heard of Xist haven't you, Nell? What does it do?
Nell - So, this is interesting. I like this because it's going back to that nice idea of, you've got your 2 X-chromosomes in a female and for that to work in a functional organism, you want to deactivate one of those because otherwise you've got two copies of every gene, everything will get a bit out of control. So, Xist is there that helps to switch off one of those X-chromosomes. It's a little piece of RNA that's doing that job.
Kat - It's the only gene that's actually made by your switched off X-chromosome, I think.
Nell - This is looking at what happens when you knock that out. So, could having two switched on copies of the X-chromosome be causing specific problems and can we find out by using this little piece of RNA to switch it off?
Kat - So, they did some really nice experiments where they actually got rid of this Xist RNA and they completely knocked out the gene that made it in mice. Then it caused problems with their blood. And actually, if you look at patients who have certain types of blood cancers, you do see problems with some of their X-chromosome genes and actually, other genes as well. So maybe they think that 'exist' might be playing a much wider role in cells. It's a really fascinating, intriguing paper.
Nell - Yeah and I like the - there was an interesting point they made at the end that some forms of leukaemia-type precursor disorders are more common in women and you can see immediately there that could be something going on there with the fact that you have got those two X-chromosomes. And if something is going wrong, meaning that perhaps parts of one are active and they shouldn't be. Perhaps that's the link there, so that's quite an interesting little trend they can see in this data.
12:34 - Can pseudogenes control cancer?
Can pseudogenes control cancer?
with Nell Barrie
Kat - And our final, kind of crazy gene RNA cancer story is about a gene called PTEN and this is a paper published in Nature Structural and Molecular Biology this week from scientists in the US. This is about pseudogenes which sound like kind of fake genes? What's going on here?
Nell - Well, this is interesting because essentially a pseudogene is a bit of DNA that's almost exactly the same sequence as a gene that's actually doing something, but it's changed slightly. And I guess probably what's happened there is it's got duplicated at some point during evolution. Because you've got two copies, you don't need both. So, an mutation in one of these copied genes isn't going to cause a problem. So you end up with a slightly mutated gene which becomes a pseudogene. Now obviously, the standard idea would be that this isn't actually doing anything. It's not coding in the right way for the gene. It's been mutated so it doesn't matter. It's just a piece of what we used to call 'junk DNA'.
Kat - But we're not allowed to say that anymore.
Nell - No, we're not allowed to say that anymore. And that's because it turns out that it could actually be doing something. So, it's not quite as simple as it first appeared.
Kat - So, what does it looks like it's doing? So, this is a pseudogene for a cancer gene called PTEN which is normally a tumour suppressor.
Nell - Yeah and I mean, the interesting thing here is that because it's so similar to PTEN, it's actually interfering with normal processes involving PTEN inside the cell. So for example, the pseudogene can actually suppress the promoter region for PTEN so essentially, that's turning it off. And it can also bind, soak up little pieces of microRNA that are meant to be interacting with PTEN. So, it's kind of subverting the normal processes by being such an almost exact copy within the cell.
Kat - I think it's really interesting putting this in the context of what we are now starting to understand about how the levels of gene activity are important. It's not so much about, "This gene is on. This gene is off." It's actually, "Do you have an extra dose? Are you making a bit more or a bit less?" And gene regulation is getting really complicated because it seems to be maybe about the subtle levels. So, perhaps that's playing a role here, but thanks very much Nell.
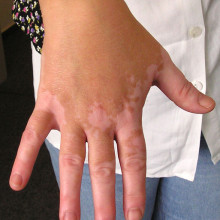
14:37 - Protein could treat vitiligo
Protein could treat vitiligo
Writing in the journal Science Translational Medicine, researchers at Loyola University Chicago have developed a modified protein that could potentially treat the skin condition vitiligo, which causes white patches on the skin and affects around one in every two hundred people worldwide. There are no really effective treatments, and therapies such as light treatment, steroid creams and skin grafts can cause significant side effects and don't prevent the disease from progressing.
The disease is caused by a person's immune system turning on their own pigment cells, which are responsible for giving skin its colour. Previous studies have shown that a protein called HSP70i is involved in this overactive immune response. The researchers created a version of HSP70i where one single building block, or amino acid, had been altered. They discovered that this mutant protein replaces the normal version in cells and switches off the immune reaction.
When they gave the mutant HSP70i to mice with vitiligo, they found that it returned their black and white fur to normal black colouring. Similar effects were seen in samples of human skin with the condition. Although the work has only been done in mice and skin samples in the lab so far, the researchers plan to run a clinical trial in patients as soon as possible.

15:54 - Reading the genome
Reading the genome
Scientists at the US Lawrence Berkeley National Laboratory have made a major step forward in understanding how the information in our DNA is "read" - a process known as transcription - publishing their results in the journal Nature.
Eva Nogales and her team used a technique called cryo-electron microscopy to zoom in on the molecular machinery responsible for transcription, taking snapshots of its structure at an incredibly detailed level. This machinery, known as the "transcription pre-initiation complex", contains a number of different proteins which help to load RNA polymerase II - the enzyme responsible for "reading" genes - onto DNA so it can start working. In these experiments, the scientists produced some of the components of the complex in a test-tube so they could study them relatively easily.
The snapshots taken by the team help to reveal how RNA polymerase II is attracted to DNA and kept there, as well as how it chooses the exact place to start reading a gene. Next, the researchers plan to work towards spying on the whole transcriptional machinery - a huge structure in molecular terms. And although it's technically challenging, this kind of research is helping us to understand how our genes work on the deepest level.

17:03 - Sea Lamprey brain disease link
Sea Lamprey brain disease link
Researchers at the US Marine Biological Laboratory have made some surprising discoveries after decoding the genome of the sea lamprey, a primitive fish, reporting their work in the journal Nature Genetics. Although they're very different from us, the sea lamprey's genome contains genes linked to human neurological conditions including Alzheimer's and Parkinson's diseases, as well as spinal cord injuries.
Unlike humans, lampreys have an incredible capacity to repair their nervous system - for example, if their spinal cord is cut, they can regenerate it and be back swimming around in as little as ten weeks. But, intriguingly, lampreys don't have myelin - the insulating material that surrounds our nerve cells. And it's the molecules associated with myelin that prevent human nerve cells from regenerating when they're damaged. But the researchers found that lampreys also have these myelin-associated molecules, yet don't actually make myelin.
By opening up the black box of the lamprey genome, scientists can now find out what's going on here, and find out whether the lamprey's regenerative abilities can be exploited for healing human nerves.

18:22 - Dr Dianne Newbury - Language disorders
Dr Dianne Newbury - Language disorders
with Dr Dianne Newbury, Wellcome Trust Centre for Human Genetics, Oxford
But now it's time to turn to the genetics of another developmental disorder - Specific Language Impairment, or SLI. Up to two children in every classroom of 5-year-olds may have this condition, meaning that they have difficulty developing their language skills. To discover more about SLI, and how genetic research is helping scientists to understand it better, I spoke to Dr Dianne Newbury at the Wellcome Trust Centre for Human Genetics in Oxford.
Dianne - So, specific language impairment is essentially just children who have problems developing and learning to use language for no apparent reason. So, they don't have problems in other developmental areas, but just have this very specific problem with learning to use language.
Kat - So, they don't have hearing problems, they don't have autism?
Dianne - No. So, speech and language problems are quite common in children who have other developmental problems, so like you say hearing problems, autism. But the children we look at don't have any of those associated problems. Their main clinical concern is their speech and language impairments. Some children just have a language delay and then they catch up. Generally, the children that we look at with SLI, they are always behind their peers in terms of language.
You can imagine, if you start school and you've already got problems with language, that's going to compound as you go through school because obviously, it's such an important factor in terms of educational attainment. But a lot of children learn to compensate for their problems so if you were talking to someone with SLI, you might not notice that they had SLI. But they report that they always have problems - they have to concentrate harder than other people when listening to language and when trying to construct sentences as well.
Kat - So, what gave you the first inklings that there might be something genetic to do with this kind of condition?
Dianne - So, when I first came and started working in Oxford, I was working on dyslexia which is kind of a similar condition, but with written language rather than spoken language. At that time, there had been very little work done on specific language impairment, but there is somebody in Oxford called Dorothy Bishop who had done a lot of work in families and with twins, and she'd been looking at the way that SLI runs in families. So there seems that there's quite a strong kind of heritable link. So, if your parents had language problems, then you're more likely to have language problems yourselves. And if you look in twins who are genetically identical, so identical twins, if one of them is affected, the other probably has a much higher chance of being affected. So, that gives us quite a good indication that there's some genetic contribution involved in the disorder. It's not kind of a mutation in a specific gene which causes the disorder, but instead we're thinking about kind of normal genetic variations between individuals. They're what we call these 'risk variations' which slightly increase your risk of having a language impairment, but there's many of them scattered right across the genome and the more of them you have, the higher your chances of developing a language impairment.
Kat - Sort of a mix and match approach.
Dianne - Yeah.
Kat - So, how do you go about tracking down some of these genes that are involved in the risk of this condition?
Dianne - So, because there's so many of them involved, and each one is likely to only have quite a small effect size, they're much harder to track down than the kind of mutations which we normally think of with genetic disorders. But we apply similar kind of techniques and what we do is we collect DNA from families who are affected by specific language impairment and we compare the DNA between siblings in the families. So, you can imagine that if a pair of siblings are affected by specific language impairment, then we can sample their genetic material and we can look for regions of chromosomes which they have both inherited the same copy of from their parents. And if we do that in one family with a pair of siblings then we can narrow it down to about 50% of the genome that might carry contributory factors. If we do that across hundreds and hundreds, and hundreds of families, we should be able to narrow it down further and further, and further.
Kat - So, what have you found so far? Are there any really good candidates you've got?
Dianne - Using that technique, we manage to identify region on chromosome 16 and a region on chromosome 19 which we think might carry genes that contribute to language impairment. And then using another method called association, we manage to narrow it down from these chromosome regions to 2 particular genes on chromosome 16. And one is called ATP2C2...
Kat - Catchy...
Dianne - And the other one is called CMIP, so yeah, very catchy names.
Kat - Do you know anything about what these genes actually do in the body?
Dianne - So, ATP2C2, there's a little bit known about it and we know that it's a calcium transporter protein and so, what it does is it pumps calcium out of the cell for transport out of the cell, and that's quite interesting because calcium regulation is known to be involved in memory kind of processes, to be quite important in neuronal processes. So, that might link somewhere back into why children have language impairment. The other gene, CMIP, there's less known about it, but we know it's involved in the scaffolding of the cells, making the shape of the cell. And that might be interesting in terms of when neurons project and connect to each other. It may be important in those kind of processes, but that's a bit less clear for that gene.
Kat - So now that you found these genes and you're looking for more, what's the hope that this knowledge could actually maybe lead to better therapies for kids affected by this disorder?
Dianne - My hope would be that it will provide us with a better understanding of why certain children are at higher risk of language impairment because given that it's quite a common disorder, it affects 5% of the child population, but we really don't understand why these children have language disorder, the differences between different children - so SLI in one child might look very different from SLI in another child and we don't really understand the best way to categorise it or what the underlying problem is.
So, if we could understand what the kinds of proteins and biological processes that are involved in it, then maybe that would help us to have a better understanding of how to categorise it and how to help the children. So, for example, if we knew that some children had variations in ATP2C2, that for those particular children, memory was important then we could use some kind of memory enhancing training in those children, and that would then help hopefully with their language impairment.
Kat - That was Dianne Newbury from the Wellcome Trust Centre for Human Genetics. And if you want to find out more about SLI, search online for the RALLI Campaign - that's R A L L I.
25:39 - Gene of the month - Fruitless
Gene of the month - Fruitless
with Kat Arney
And finally, our gene of the month is Fruitless. Although it sounds like a bit of a lost cause, the Fruitless gene has attracted plenty of interest - not to mention controversy - over the years. In purely biochemical terms, Fruitless encodes a type of protein known as a 'transcription factor', which can switch other genes on, but it's only when you look at its biological role that things get really interesting, because male fruit flies with faulty Fruitless have problems getting down to mating with female flies - in fact, they don't seem very interested in the ladies at all, with some mutants preferring to go for the boys. And female flies with faulty Fruitless tend to behave more like males. Fruitless also pops up in mosquitoes and other insects, where it's involved in sex determination
In fact, the gene's name was originally "Fruity" - a slang term for gay - but it was switched as public attitudes to homosexuality became more enlightened. But while much has been made of Fruitless in the media, with speculation about whether the rules of human love and sexual orientation can be inferred from fruit fly genetics, humans don't actually have an obvious version of the gene. So that kind of conjecture is probably as fruitless as the gene itself.
25:39 - Is there a cheap reliable way to predict protein structure from DNA?
Is there a cheap reliable way to predict protein structure from DNA?
Stephen Serjeant asks “Is there a cheap, easy & reliable way of predicting protein structure from DNA or protein sequence please?”
We asked Dr Tina Perica, from the MRC Laboratory of Molecular Biology in Cambridge...
Tina - The short answer to the question, how easy it is to predict the structure from sequence is, sometimes it’s very easy and sometimes it’s very, very hard. Proteins in cells of all living organisms are linear polymers of about 20 different building blocks which we call amino acids and the way they fold into their 3D structures is what determines their function. So, if a protein is able to fold to a single and specific 3D structure, us scientists should in theory also be able to predict that same structure by just looking at its sequence.
It is easy if there is a homologous protein for which the 3D structure is already determined, for example by x-ray crystallography. Homologous means that proteins are evolutionarily related. It means that they are protein brothers or cousins. So, if we know the 3D structure of a closely related protein, we can easily and quite accurately predict what the structure of its close cousin will be. What can we do if we can't find any protein cousins? Well then, it gets harder. But luckily, although only in the human genome, we have around 25,000 different proteins, we have so far in all of nature, found only a couple of thousand different ways in which any protein can fold. So, we can then take our sequence string and try to thread it in all of those possible combinations.
Every year, there are more and more new experimentally solved protein structures, but there are fewer and fewer new folds. However now and again, we do have a completely new protein which folds in a way we have never seen before. And here, we come to the very, very, very hard predictions. In these cases, there is nothing else to do, but to use all of our knowledge in biophysics and chemistry and just start from scratch.
Kat - That was Dr. Tina Perica, at the MRC LMB in Cambridge.
Related Content
- Previous Why does Washing Shrink?
- Next John Snow and Cholera









Comments
Add a comment