Genetically modified, or GM, crops are a hot topic. Some people are deeply suspicious of the technology while others see it as an effective and efficient way of generating bountiful, healthier harvests. Plus, purple tomatoes, a giant of a gene involved in heart disease, and what's in a name? We take a look at the naming of genes.
In this episode

01:02 - Jonathan Jones - GM crops
Jonathan Jones - GM crops
with Jonathan Jones, The Sainsbury Laboratory
Kat - Few scientific topics are as controversial as GM technology, used to manipulate the genes of food and other crops to make them resistant to diseases and pests, tolerate poor growing conditions, or to increase their nutritional value. To take a look at the science of GM behind the hype and headlines, I spoke to Professor Jonathan Jones at the Sainsbury Laboratory in Norwich.
Jonathan - GM is actually a method, not a thing. It's very important to bear that in mind and the method enables you to take DNA sequence from essentially any organism and use the properties of a bacterium called agrobacterium to deliver that DNA into a plant cell. If that DNA carries a gene that serves a useful purpose, for example enhances crop resistance to insects, then you can get a plant back that has properties that you could not have achieved by plant breeding.
Kat - So, can you give me some examples of this? What sort of properties have been given to plants from the outside by this technology?
Jonathan - Well, two most abundantly used examples are firstly to use a gene from a bacterium called Bacillus thuringiensis which is often used by organic farmers. It contains a so-called crystal protein that's toxic to larvae of many insects. The plant is engineered to carry in its cells the gene that makes these proteins. So, the protein is made in the plant and anything that eats the plant that is susceptible to that protein doesn't thrive or dies whereas insects - and this is very important - that don't eat the plant are completely unaffected. And so, this is much better as a way to control insect pests than applying insecticides from sprays or airplanes or combines or whatever, because you get much less collateral damage to non-target insects. It only kills the insects that eats the plant. Approximately, 400,000 tons of insecticides - we're talking nasty neurotoxins - have not been applied that would otherwise have been applied to control insects. And that's an enormous benefit that's built up over the last 20 years of cultivation of GM crops.
Kat - So, this all sounds like a good thing in theory, but is there any, for example, if you have a crop that's been engineered, to have a bacterial toxin in, I'm going to be eating that crop if they make any food from it. Is there any risk to human health from that?
Jonathan - Well, with respect to Bt, like I say, organic farmers apply Bt. There's no credible mechanism for any hazard to human health. There's never been any evidence that it causes any damage to human health. So no, not a problem.
Kat - What are some of the things that scientists are now working on now, these kind of characteristics that we could engineer into crops?
Jonathan - The scope for improvement of crops in many, many different ways, some more near-term and some more distant. Near-term - some of your listeners may have heard about a trial of GM Camelina which is engineered to produce the long chain polyunsaturated fatty acids that are present in fish oil and very good for vascular health. There's just not enough fish in the sea for us all to get enough of this stuff. These compounds are only made in algae that were eaten by little invertebrates that were eaten by little fish, that were eaten by big fish that we eat. But now, it's possible to make these compounds in an oil seed. This would greatly improve the environmental sustainability of fish farming and increase the supply of these compounds in our diet. So, that's a good thing. My own work, we're working on potato late blight resistance as other research is elsewhere. Potatoes are sprayed 15 times a year for late blight. It is expensive increase of cost in production and potentially damaging, tractors going up and down, CO2 costs, and so on. So, if we can make the plant resistant to disease by moving in genes wild relatives of potato into the cultivated potato then we could save a lot of environmental impact. And that same principles can be applied to many other diseases of many other crops. So longer term - there are credible approaches to increase drought tolerance, to increase salt tolerance, and farthest off in the future of all, trying to engineer nitrogen-fixing versions of crops like wheat and barley, and maize.
Kat - So, you wouldn't need to add fertilisers. They kind of make their own fertilisers from their roots.
Jonathan - That's right. So, if you look in Africa, one of the major constraints on crop yields is lack of availability of nitrogen. There's hardly any fertiliser factories in Africa. So, if you could get the plants to fix nitrogen from the air themselves without any need to supply exogenous fertiliser, that would be a good thing.
Kat - GM technology is one way of getting genes into things, of changing the genes of an organism. Could the kind of changes you're talking about not be achieved in other ways? Why is GM (genetic modification) the only way to do some of these things?
Jonathan - Plant breeding is really great. Plant breeding is going on. Plant breeding has improved in efficiency enormously by the advent of new sequencing technologies - to suggest it's a matter of either GM or plant breeding is a completely false antithesis. So, both have contributions to make. The problem with just breeding them in, as you breed them in one at a time, if you've only got one resistance gene, it's easy for one mutation in the pathogen to overcome that resistance. Also, when you bring in genes by breeding, you bring in big chunks of chromosome from the wild relative, not just the gene you want. Compare, if you will, a scenario where you clone multiple resistant genes from different wild relatives. And then you got the cloned DNA, you can stick them together so you can put in 3 genes at once. You leave behind all the other bad alleles of other genes that actually decrease yield and that is a much better way to give you sustainable disease resistance than bringing in these genes by breeding. Particular benefit is, once you've got 3 genes stuck together by recombinant DNA then they can't be broken up by plant breeders whereas if you bred in 3 different genes and they're in different places in the chromosome, of course, once you got a variety, a plant breeder will start crossing from that resistant variety into their own they will separate all those genes and then they can be picked off by the pathogen. But once they're ligated together, they can't be separated. So, that's a good illustration of why recombinant DNA is better than plant breeding in this particular example. But also, why it's very important to preserve genetic diversity so we've got biggest possible resource of diversity to mine for new sources of resistance to important diseases.
Kat - If you look on something like social media, often there are criticisms of some of the big agricultural companies that are doing genetic modification. And there seems to be a fear or the idea that it's bad or risky, or that by involving big businesses in this, it's somehow suppressing farmers or it is a bad thing for the world. How would you answer some of those criticisms?
Jonathan - I think that the technology is usually a surrogate for people's other concerns. You know, the technology becoming a lightning rod for people's legitimate questions about, is it a good thing for only a few companies to control the germplasm on which this world's food supply depends? That's an important issue. I wouldn't contest anybody who's concerned about that issue, but it's nothing to do with GM. The right discussion to have is, what kind of agriculture do we want? What kind of genes do we want to put in? Are we putting in the right genes? Could we make better choices about how they are deployed, and so on and so forth? We actually have an urban population that is profoundly ignorant of how hard it is to produce food. The real difficulty is, that in the media, everyone's attention span, including journalists is so short you can't really develop a complex argument. Everybody just wants to deal in very simple sound bites that actually are gross oversimplifications of the problem or the solution.
Kat - That was Jonathan Jones from the Sainsbury Laboratory in Norwich.

09:14 - Cathie Martin - Purple tomatoes
Cathie Martin - Purple tomatoes
with Cathie Martin, John Innes Centre
Kat - While I was in Norwich, I also caught up with Professor Cathie Martin at the John Innes Centre. She describes herself as a metabolic engineer, using GM technology to develop fruit and veg that are even more healthy- including her famous purple tomatoes. I started by asking what exactly is metabolic engineering?
Cathie - So, it's engineering metabolism and I'm very interested in engineering a group of compounds which are called polyphenols, and those are fairly recently recognised to have health promoting effects when you eat them.
Kat - So, this is basically making fruit and veg even more healthy than it was.
Cathie - That's right. I mean, I'm a bit fed up of all of the information that you get about food being things that are bad for you. It's got too much salt, it's got too much saturated fat, etc. So, I think we need to know whether there are any things that are good in foods for us. And all of the evidence suggests that eating fruit and vegetables is very good for you. There are specific compounds in fruit and vegetables than can really reduce the risk of what we call 'chronic disease' like certain cancers, cardiovascular and metabolic disease like type II diabetes.
Kat - In front of us on a table, we've got some beautiful tomatoes, but there's two red tomatoes that look like tomatoes and then there's two yellow tomatoes, and then these 4 black plum-coloured things. What have we got here? Tell me about these?
Cathie - Okay, so this is a control. So, this is a regular tomato, well-spotted, and then this is a tomato that has been metabolically engineered to produce compounds which are called flavonols.
Kat - That's the yellow one here.
Cathie - Yeah, it's the yellow one. And the reason it's yellowish or orangey yellow is because it has high levels of these pale yellow compounds. So, if I was to squash it, you'd see a pale yellow juice which would be more yellow than in this one. And so, it's not because it's got less of the red colour which is lycopene, but it's got more of a pale yellow compound.
Kat - So, it's an extra yellow rather than not red.
Cathie - An extra yellow, yeah and these are things like quercetin and campherol which are good for you. They're good anti-oxidants, but they're also shown to beneficial in the diet.
Kat - What about these purply ones? These are gorgeous.
Cathie - These ones have another compound. It's chemically related to the flavonols but these are anthocyanins. And so, there are pigments in plants. They're the sort of things that make delphinium blue.
Kat - That looks like blackberry colour. These are blackberry coloured tomatoes.
Cathie - Yes and that, it's the same compound that's present in blackberries and all of those super fruits that you've been hearing that are good for you. So now, we've done it in tomatoes. They also have this red lycopene, but you can't really see it because they've got so much anthocyanin and what will make you gasp...
Kat - Okay, we're cutting them open. Wow!
Cathie - Isn't it beautiful?
Kat - It's like a tomato but really beautiful rich, dark purple. Literally, like a tomato, but you're looking at it through a completely purple filter, almost black as night.
Cathie - They really are tomatoes, but they look completely different because they're very rich in these anthocyanins. Actually, this one is rich in both anthocyanins and the flavonols as well and it gives this rather beautiful royal purple. But we've shown experimentally in what we call pre-clinical studies that these can slow down the rate of progression in cancer in animal models of that disease. And also, that they can be protective against arteriosclerosis when they're eaten in their diet.
Kat - But I could get those benefits from eating loads and loads of blackberries.
Cathie - Absolutely, but probably, if you eat loads and loads of blackberries, enough that you'd get say, the equivalent of 2 of these tomatoes would be about 70 grams of blackberries.
Kat - That's a lot of blackberries.
Cathie - It's a lot of blackberries. You wouldn't get them all-year-round and you'd eat them with a lot of sugar which would be a bad thing. So these, you can eat without the sugar.
Kat - Am I allowed to taste one? These are laboratory specimens.
Cathie - I think we'll have to say that they're laboratory specimens. There's a problem if you eat the seeds because we haven't got a regulatory approval and it would be considered an inadvertent environmental release and I'd have to ask personal questions.
Kat - Okay. I don't want to inadvertently release anything into the environment. These tomatoes, they look beautiful, they've got useful compounds. What other kind of traits do you think you could engineer into our fruit and veg that would be beneficial?
Cathie - Well, we've already done some other polyphenols which you may have heard of, something called resveratrol that's present in grape juice and in red wine. We're looking to be able to regulate the production of this red compound here, lycopene. We haven't got the tools yet to do it, but we're looking to make high lycopene tomatoes or even higher lycopene tomatoes.
Kat - Super tomatoes!
Cathie - Super tomatoes, because lycopene is quite healthy for you and protects against some cardiovascular disease. So, I think that the prospects, the potential is enormous, but we're just at the beginning at the moment.
Kat - How have you made these purple and yellow tomatoes? What's the process you go through to turn a red tomato into a purple tomato?
Cathie - Okay, so the genes that we use to do the engineering are essentially switches and they switch on the production of these compounds in a place where they're not normally made. So, the same compounds will be made by tomatoes in the leaves when they're stressed. And if you ever forget to water your tomato plants, you'll see these purple pigment forming at quite low levels in their leaves. What we've done is be able to switch them on by putting in a gene that serves as a molecular switch to turn on the pathway. So basically, we just moved where a metabolite, a compound is produced from the leaves to the fruit. There are actually some wild species of tomato that makes some anthocyanins in their skin of the fruit, but they don't make it on the inside so they don't have such high levels as these.
Kat - The examples of GM technology that we've talked about seem very beneficial, making our foods healthier and other types of technology, making foods more sustainable or growing with less chemicals and all these kind of things. But there are some people who say that we shouldn't be messing with our food in this way. Are there any risks of this kind of technology?
Cathie - I think that the procedure of genetic modification per se is an established neutral technology. So, that means that people have been using it in the pharmaceutical industry for the past 30, 40 years. Many of our medicines that we get now have been produced through genetic modification of microorganisms. And for modification of crops, the risks in terms of the safety of the food that you produce are only dependent on the trait that you engineer. I believe that it's possible to engineer something that's detrimental. I think that our tomatoes have proven health benefits. So, the technology is neutral. The trait itself has to be examined and regulated. I think that we have good evidence that this is a beneficial trait. For people that don't want GM products, that's okay. You don't have to buy them. But let everybody else have the choice.
Kat - Cathie Martin, from the John Innes Centre in Norwich.
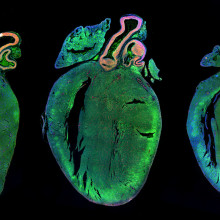
16:46 - James Ware - Tackling Titin
James Ware - Tackling Titin
with James Ware, Imperial College London
Kat - And now it's time for a look at one of the genetics stories that hit the news this month, about a condition known as dilated cardiomyopathy, where the heart muscles become enlarged and don't work properly. It affects around one in every 250 people in the UK, and is a major cause of sudden death in apparently fit and healthy individuals, as well as being the leading reason for needing a heart transplant.
In 2012, researchers discovered that one in four cases of dilated cardiomyopathy are due to faults in a huge gene called Titin. But it turns out that many people carry faults in Titin, but don't seem to have the disease. So are they at risk? I spoke to James Ware from Imperial College London, who's been looking at the Titin gene in thousands of people, to find out.
James - Titin produces the largest protein in the human body. It's a massive gene which was essentially previously too big really to study in large numbers of people. So, that really changed the landscape of the genetics of this condition.
Kat - How did you then set about seeing how faults in the gene were related to this particular heart problem?
James - It was exciting to find one in 4 cases might be explained. We were less excited to find that about one in 50 people in the general population also have a similar variant. So really, the biggest question in our mind was, why do many healthy people have these variants and how are they different from the variants that are causing disease.
Kat - So, people like you, me, regular people could be walking around with a fault in this gene that in some cases, causes a heart problem but in most cases, just doesn't.
James - It's a change in the gene that looks like it should cause a fault in a gene, if you like. What we don't know is, is it actually causing a fault in the gene but that just doesn't harm you. Is it that, it could harm you but it hasn't yet? There are lots of possibilities and that was the first thing we really wanted to disentangle.
Kat - I guess that's quite an important question because if someone's walking around with a gene fault, that means they could just drop dead at any minute, you would want to know about it.
James - Exactly. It's a huge question. Particularly, as your listeners may have heard press about things like Hundred Thousand Genomes project, even the prime minister saying he's going to sequence the genomes of 100,000 people. Well, a thousand of them will have a variant of this sort in Titin. Do we need to get them all into cardiology clinics urgently? Do we need to tell them all that their families are at risk? More and more people having genome sequencing for other reasons when they're healthy to start with. So, we really need to understand that.
Kat - So, how did you go about then trying to unpick what the faults in Titin might mean for someone who carries one of these mistakes?
James - We looked at more than 5,000 people from a whole of backgrounds. So, we sequenced some people who have dilated cardiomyopathy who were severely ill and were waiting for transplant. We sequenced people who just came to our clinic for a heart scan with dilated cardiomyopathy, and then we sequenced many thousands of people who had just been selected from the community. They were apparently healthy. And so, in all of those people, we sequenced this gene and we catalogued all the variation that we saw. We also had some heart tissue samples from people who'd consented to give us a sample at the time of transplantation. And so, we were able to look in detail how the Titin molecule had changed in people with and without abnormalities in the protein.
Kat - So, what's the bottom line? What did you find?
James - The bottom line is, firstly, that we were able to find features that discriminated between the variants that cause disease and those that don't. So, we can categorise certain types of variants. We can now reassure people that is not a variant that causes a problem. If we find it incidentally, we can reassure them we won't drag them into a cardiology clinic and that's fantastic news for them. Secondly, there are other variants that we now can be very confident they are causing trouble. The most relevant use of this is, when we find an individual who has dilated cardiomyopathy, we want to find out whether their family members are also at risk.
We can do that by examining them in the clinic and doing a scan of their heart. But very often, that would be normal. We don't know if it's simply that they haven't got the condition yet and we need to keep an eye on them. And so, what happens at the moment is people, a brother, a sister, a father, a child of someone with dilated cardiomyopathy will probably be followed in a cardiology clinic for the rest of their life. What we can now do is test their gene, test their Titin gene in the person who has the condition in the family. If we can pinpoint the exact genetic cause, we can then test the family for that and anyone who doesn't have it can be reassured, they can be discharged from the clinic. They don't have to have that long term follow up. That's a big burden for them and also for the NHS. So, I think that's the biggest of win from this study.
Kat - As we start sequencing more and more genomes from more and more people, it's becoming clear that there are these bad variations popping up all the time in people who don't appear to be ill. Do you think the work that you've done might be almost a flagship research project for this kind of investigation for other conditions? Are there any other diseases you know about where this kind of thing might be relevant?
James - Definitely. I think that one of the things we need to do at the moment is to better understand the variation in normal people and how we can distinguish between variants that are going to cause trouble in the future and variants that aren't. I think we need two things for that. One is we need more knowledge of the genetic variation present in a population. The second part is really knowing the precise phenotype, the clinical situation of the people who have those variants. That is more challenging because although we can sequence people from the general population, we can't get everyone back to do heart scans or brain scans, or whatever test is appropriate for the particular condition. So, that is the next phase I think. And projects like the Hundred Thousand Genomes project in the UK are really going to help to catalogue the clinical situation of the people whose sequences we are starting to look at.
Kat - That was James Ware, from Imperial College London. That work was published in the journal Science Translational Medicine, and we'll be taking a closer look at the Hundred Thousand Genomes project he mentioned in next month's podcast.

What are the rules about naming genes?
Kat - At this point we'd usually be hearing about our gene of the month - over the past couple of years we've had some wonderfully-name examples, from Sonic Hedgehog to Superman. Most of these are named by researchers working with model organisms such as fruit flies, who tend to pick a name inspired by the appearance of a fly lacking that gene. But listener Nicky Peng wants to know more and has written in asking: "Is there a classification system and agreed nomenclature for genes (as there are for plants and animals) or can people who discover them call them whatever they like?"
I spoke to Elspeth Bruford, who leads the HUGO Gene Nomenclature Committee - they're the people who get to decide the names of human genes - to find out what's in a gene's name, and how they pick them.
Elspeth - Obviously, we take into account if a gene has been published. We try to discuss with the authors who have published on that gene. If a gene hasn't been reported in any scientific publications then really, it's up to us and we have to look at features of the gene and decide how we think it should best be named.
Kat - Now, some of the genes that I've featured in the podcast have wonderful names because they were discovered in things like fruit flies. So, you have genes like Eyeless or Wingless and all these kind of things. Those aren't the kind of names that come through as human gene names when the equivalent gene is found in humans, are they?
Elspeth - Sometimes they are but then really, we try not to use the more whimsical names that some fruit fly researchers have chosen to use. Mainly because the aim of human gene symbols is that they should be used in all context. So, not just in scientific publications or presentations or discussions, but also - and you see this more and more - that our gene names should be used in the media, they should be used by clinicians in discussions with patients and by GPs. And especially if you're in a scenario where you're telling a patient or the family of a patient with a hereditary disorder that their child for example has a mutation in a specific gene, if the name is too comical, it's really not appropriate in that setting and some people can find it actually offensive or distressing. So, we have to take really maybe more of a wider context into account when we're naming humans genes. For fruit fly researchers, it's mainly about what's discussed in a scientific context and in the lab perhaps. But for us, we have to look at the wider picture because obviously, human genes impact human health.
Kat - When it comes to naming human genes, what sort of names do you choose and how do you reflect what the gene is like or what it does?
Elspeth - Okay, so we have a few criteria that we run through. Ideally, we would name a human gene based on a known function of the gene product. So for example, if the gene encodes for an enzyme with a known function then we would try and name it on the basis of that enzyme. If there's not a known function then we would start looking at homology. So, how related it is to other genes that are already known or maybe have a known function. As in the case of genes that are related to fruit fly genes, we would look at homologues in other species and see what was known about their functions. If there's nothing like that or not a high level of homology then we would start looking at the structure of the protein encoded by the gene. Does it encode any specific regions, usually called protein domains or motifs? And then we would perhaps bring that into the naming. And we'll also of course take into account any information that has been published or any information that a researcher has come to us saying, "We know something about this gene or this gene product." So really, a variety of criteria but our favourite criteria is to name based on a known function of the encoded gene product.
Kat - This is going to be a strange question but do you have any particular favourite gene names or gene names that really stick out for you?
Elspeth - You get fond of the ones that you remember naming! So, one way we like to name genes is grouping them into gene families. A gene family is usually related by sequence similarities. So, the genes are all related to each other and as a result, they quite often have related functions, not necessarily the same function, but something similar about their function. It's quite rewarding to take a group of genes that have been named very disperately and then contact lots of lots of researchers - because that is the key part of our job is actually contacting the researchers because obviously, they know a lot more about these genes and the encoded proteins than we do. So, we contact researchers and then we say, "This gene is a member of the family along with these other genes and we'd like to group them altogether and name them like that so that other researchers can pull out the whole family." And so, there's a few gene families that I've worked on in the past that it's quite rewarding then when you see them being published and people are using the new nomenclature. What's most important for us is that whatever name we decide upon will be used, because if the name is not used then it's kind of defeating the purpose. And the purpose is for everybody to be able to find a name in the literature in publications and then know exactly which gene somebody is talking about.
Kat - Do you think we will see a day when all the whimsical names will be replaced by three-letter acronyms?
Elspeth - Well certainly, for human - I don't know if all of them because some of them are not offensive or pejorative in any way. I don't know if all of them will go. I mean, Sonic Hedgehog - sometimes people say that Sonic Hedgehog has been replaced. It hasn't. I mean, Sonic Hedgehog is just so entrenched in the literature that I think we'd be kind of cutting off our nose to spite our face to get rid of it. But some of the other ones like Lunatic Fringe or something. Yeah, when it's actually potentially offensive, we really have to get rid of them. I think they'll probably stick around in Drosophila though because they don't see any reason to change them and in fact, they think our way of naming is very boring, too prosaic, but we like to think it's more informative. Less fun perhaps!
Kat - Elspeth Bruford from the HUGO Gene Nomenclature Committee.
- Previous Space Worms
- Next Detecting dark matter









Comments
Add a comment