elife Episode 2: Plants keep Thyme, cancer drug resistance and clear corneas
How plants do molecular mathematics to "thyme" their starch consumption, how cancers evolve resistance to chemotherapy and how to combat it, how the retina and cornea keep themselves clear of blood vessels, watching T cell receptors in real time and do governments listen to scientists?
In this episode

00:44 - Plants count in their sleep
Plants count in their sleep
with Alison Smith, Martin Howard, John Innes Centre
Plants use molecular mathematics through the night to stave off starvation! Alison Smith and Martin Howard from the John Innes Centre explain the problem posed to plants when the sun dips to Naked Scientist Martha Henriques...
Martin - During the night, the sun goes down. That's a pretty fundamental problem here, in that during the day, plants can of course get energy from sunlight through photosynthesis. But during the night, when the sun goes down, their source of energy disappears and they had to come up with some way to compensate for that so the plant metabolism and growth can go on during the night. So, the way in which plants solve this problem is that during the day, they store some of the energy which they get from sunlight in the form of starch and during the night, this starch is broken down. And the observation was that the rate at which starch was broken down during the night was more or less constant during the night. If you measure the amount of starch as a function of time during the night, it kind of goes down as a straight line.
Martha - Okay, so it seems that the plants are budgeting correctly for the amount of daylight that there is.
Martin - That's right. They somehow seem to know how much starch they have and they know how much time there is left until dawn, and then they budget appropriately for the starch degradation rate based on those two numbers. Interestingly, if you give the plants unexpectedly early night, so these plants which we have in the growth rooms have spent their entire lives in one light regime and suddenly you give them an earlier night than they've ever seen before, they're able to seamlessly re-adjust, rebudget so that they can adjust their starch degradation, such that they ran out of their food reserves exactly at the time of expected dawn - even when the sunlight has disappeared earlier than they were expecting.
Martha - And so, your new interpretation in this study is that the plants were actually dividing one quantity by another. How did you come up with this idea?
Martin - Well, that seemed to be the only hypothesis that was consistent with our data. It didn't seem to matter how we tried to mess up the plants and the sunlight regimes to which they're exposed, whiether we gave them early nights, late nights, whether we altered the intensity of light during the day. We tried lots of things to try to interfere with this process but whatever we did, the plants seem to be able to rebudget appropriately. They would always get the right rates of degradation during the night, so they'd run out of their food reserves at dawn. The only way this seems to be consistent with our data and to be able to do that in all circumstances, was that they were really doing this division calculation, that they knew, that plants knew how much starch they have. They'll have information about how much time there was to dawn and they would divide these two numbers together to work out the right rate to eat up their starch.
Martha - And what exactly does it mean for a plant to divide one number by another as plants don't have a brain to do calculations? What does it mean for a plant to do arithmetic division on a molecular level?
Martin - Right. I mean, it does sound rather an extraordinary thing. We were certainly struck by this interpretation. The way in which we think it doesn't work, thinking about it naively, could it be like a computer or a calculator? Is a plant doing a calculation like it's done on a computer? And we think that's extremely unlikely because the way in which computers do these calculations is very, very complicated. We think it's much more likely that it's implemented in a completely separate way. There are two molecules that are giving the plant information about the amount of starch and the time to dawn. Basically, one of these molecules is enhancing the rate of starch degradation and the other molecule which is measuring the time to dawn is basically inhibiting it. So, if you have one molecule that's trying to make something happen, another molecule that's trying to stop it, that's a little bit like doing a division calculation because the more you have the molecule that's trying to stop it, the slower the rates you have, and that's rather like doing a division calculation. So, it turns out that if you have these two molecules and you set up the right set of chemical reactions then you can create a degradation rate that's the ratio of these two quantities. That's how we think this is done. It's not really done digitally like it is in your computer. It's done in a much more analogue fashion through chemical reactions.
Martha - And part of this analogue reaction is regulated by the internal clock that the plants have.
Martin - Yeah, so the molecule that's giving the information about the time to dawn comes from the circadian clock. So, plants just like us have an internal circadian clock which tells them when dawn is going to come and when the sun is going to rise so they can switch on the appropriate genes to do the right things at that time. So, we think that the timing information comes from the circadian clock and indeed, I think we're fairly certain about that because we know that in certain plants where the circadian clock has been interfered with, the plants start to make decisions that are incorrect.
Martha - And Alison, this work happened through quite a novel collaboration and came up with a very novel result.
Alison - The idea of any organism doing arithmetic division is new. This may be something that needs to be looked at more widely in different types of organisms. We're very surprised and excited by this, the fact that plants can do maths and they sit there in the night and they have to do maths. So, this is perhaps something else that I'd like to emphasise. This is not some trivial thing. If the plant gets the sum wrong, so it degrades its starch too fast and runs out of starch before dawn, we know that the plant gets into serious problems. We have mutants which do this and those plants grow more slowly because they can't manage their carbon budgets properly. So, this has profound implications for plant productivity, obviously, with knock-on effects on the way that we might study how to increase plant productivity in the future.
Similarly, if the plant gets the sun wrong and doesn't use all of its starch up, then it's wasting energy that it stored. Again, it's less productive. So, this is really important for plant productivity. Just as a very rough back-of-an-envelope calculation, perhaps a third of the carbon that's in life on Earth went through this metabolic pathway. So understanding how the plant controls this is really quite fundamental to biology in general.
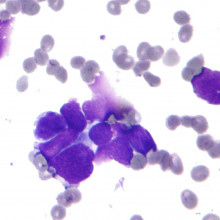
06:58 - Evolution of cancer drug resistance
Evolution of cancer drug resistance
with Martin Nowak, Professor of Maths and Biology, Harvard
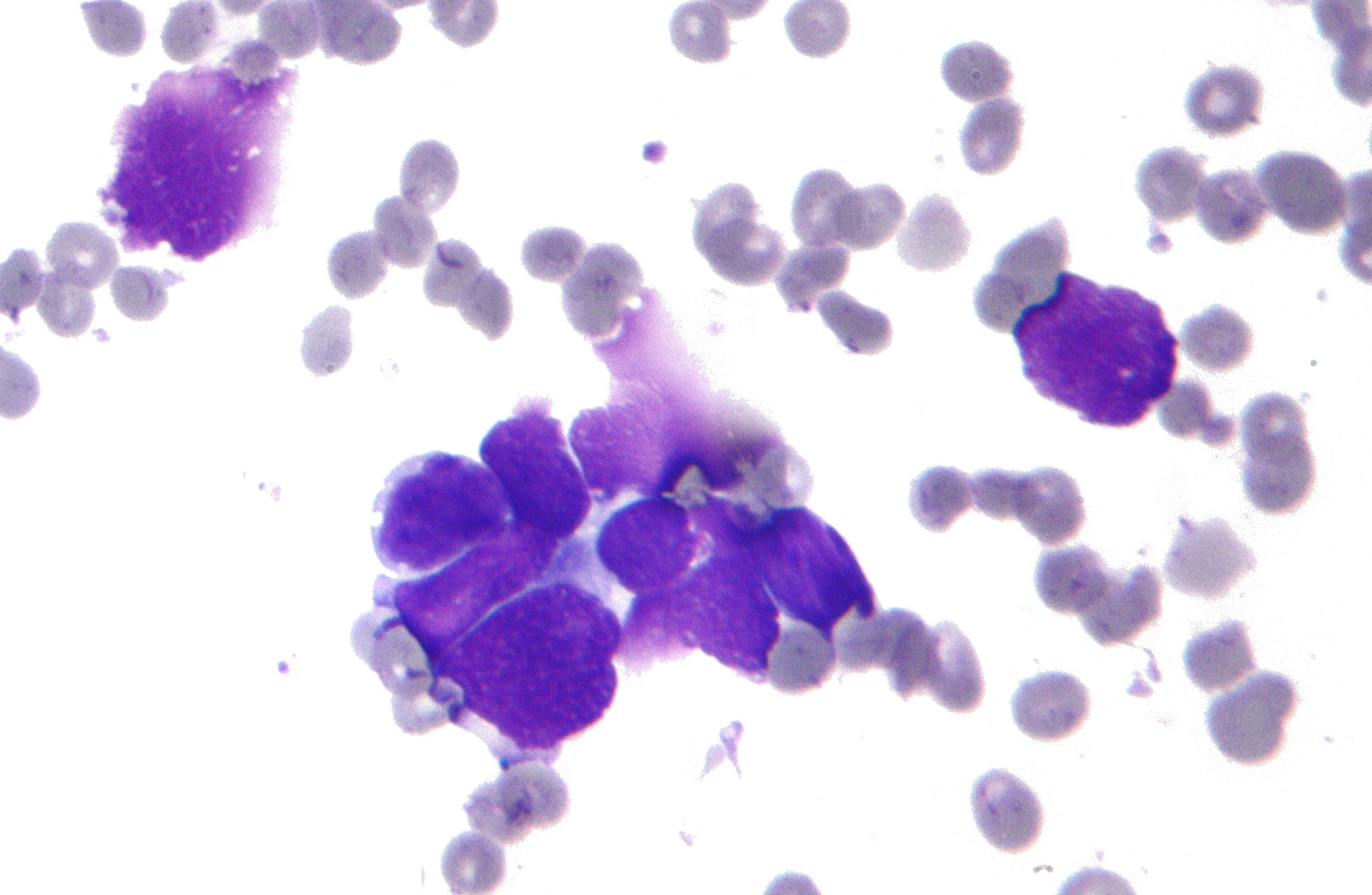 Cancer researchers and doctors all over the world are getting increasingly excited about the new era of molecular therapies designed to target specific gene faults in tumours. But while these drugs tend to show impressive results, their effectiveness soon wanes and the cancer comes back after a number of months or years, as it evolves resistance to the therapy.
Cancer researchers and doctors all over the world are getting increasingly excited about the new era of molecular therapies designed to target specific gene faults in tumours. But while these drugs tend to show impressive results, their effectiveness soon wanes and the cancer comes back after a number of months or years, as it evolves resistance to the therapy.
Kat Arney spoke to Martin Nowak, Professor of Mathematics and Biology at Harvard University, who has been using mathematical models of tumour evolution to try and understand why this happens. His results could change the way that these new treatments are used.
Martin - The initial effect of the drug is often amazing and doctors see a decline in advanced cancers to an extent that they have never witnessed before in any situation. So, this is extremely promising, but many of these therapies exactly as you say, they are short-lived and the reason is because the cancer cells develop resistance.
Kat - They kind of evolve don't they and change to escape the drug.
Martin - Yes, exactly. So, we are witnessing here an evolutionary process on a pretty fast timescale and here's also where the mathematics comes in because the mathematics of biology is very developed to understand evolution. So, evolutionary dynamics can be described with exact mathematical equations. We can describe how populations get a resistance, get mutations and then how mutations respond to selection pressure. This is what we're doing for cancer here.
Kat - How do you go about starting to model what seems like such a complex biological process?
Martin - The first question would be an estimate of how big the population of cancer cells in the body is. So, if you look at the particular lesion of a certain size then very often we're confronted with approximately 1 billion cancer cells. And then we give the treatment and the observations suggest that there are single point mutations that can actually confer resistance to the treatment. So the first question that would arise is; at the time to start treatment, are the resistant mutations already present in the patient, or do the resistant mutations emerge once therapy has started.
Kat - So, that would be single changes to a gene that would make the cells resistant to a treatment or not, to find out whether they're there at the start or whether they kind of come on later.
Martin - Yes, that's exactly right. So now, the question is; supposed, there are a number of point mutations in the oncogene that can do this between say, 1 and 10 or so. The point mutational rate is fairly well known approximately 10 to the minus 9 per base per cell division. So, given this point mutation rate, given the population size, we can fairly accurately estimate the probability that the resistant cells are already there before you started treatment.
Kat - So, by doing some maths, you can kind of figure out the rate at which these cells are mutating and evolving, and kind of track back to work out whether they were there at the start.
Martin - Yes, exactly. So, if you are telling me basically there are 10 possible point mutations, the population start is 10 to the 9, the point mutation is 10 to the minus 9 then there's some very high probability that we can conclude that the resistant mutations are already there by the time the doctor starts the treatment. So maybe 1 in a million cells has the resistant mutation and this is below clinical detection. So, if you examine the cancer, you would conclude that there's no resistant cell present because the frequency is so small. But then you start treatment and within approximately 20 to 30 weeks, you observe that the treatment fails. If you now look and analyse the genetic composition of the oncogene, you will find that the resistant mutations have emerged.
Kat - This seems to me to be incredibly significant for treatment because at the moment, with these targeted treatments, we tend to test one at a time. So, you do an analysis of a patient's tumour. You say, "Okay, you need this drug" and then you give it to them for a bit, it works and then it fails, and then you try another one. What conclusions can you draw from your research about how we should use these drugs now?
Martin - So, what we have done in this paper is really to ask the question, supposed that you are in a situation where you have two targeted therapies against the same cancer. If you now lose two therapies at the same time then what is the probability that the cancer is now cured. This really depends on whether or not there exist single point mutations in the genome that give simultaneous resistance to both drugs. So, if you are in a situation where there is even only a single point mutation in the whole genome that confers resistance to both drugs at the same time, the situation again is very gloomy and the chances for curing patients with this type of combination therapy are fairly small. So, what we really need to design are drugs that work against the same cancer and moreover, there's not a single point mutation in the genome that keeps rise to resistance to both drugs at the same time. Then the cancer has to respond by developing at least two-point mutations. One that gives resistance for drug A, one for drug B, and that is not so easy for the cancer.
Kat - Cancer cells only have a limited range of ways they can get around these drugs. They're not completely immortal so you would think that eventually, if we had enough targeted drugs, we would be able to have enough ways to hit cancer so it couldn't get round the treatment.
Martin - Yes, this would be the idea, but always keep in mind, it doesn't matter how many drugs you have, if the cancer cell has a mechanism to give resistance to all those drugs simultaneously. So, you really must develop drugs that do not have resistance mutations in common and here the surprising observation of our current study is that a single point mutation of the genome that would give you resistance to both drugs would defeat the treatment. The very important aspect is that whatever combination therapy you use, do not use it sequentially because to use first one drug and then wait until it fails, and then another drug is a certain recipie for complete treatment failure. To use two drugs simultaneously as opposed to sequentially is a much better idea and this is rare in the clinical practice of cancer treatment at the moment.
Kat - How quickly do you think you can get this message out to people who are treating cancer patients and for example, running clinical trials of these new targeted drugs? It seems like an incredibly important thing for doctors to know about.
Martin - Yes, I think it can happen quickly. I was involved in a similar study in 1995 for HIV virus and there, we actually were able to show that single drug therapy of the HIV virus gives rise to complete resistance in approximately 4 weeks depending on the drug because there's a very rapid turnover of the virus. This moved the field very quickly, I mean within in a year to combination therapy. I mean, for me, it's fascinating to see and to hope that mathematics can really help to inform experimental scientists and doctors in the clinic. And so, my hope would be that in a few decades from now, our understanding of cancer will be engineer-like and then we can really use an engineering-like approach to actually design and use the optimum therapies and prevent many, many people from dying from cancer.
14:50 - Seeing Red - Blood vessels in the eye
Seeing Red - Blood vessels in the eye
with Bala Ambati, University of Utah
There are at least two places in the body where blood vessels are actively  discouraged from growing: one is the cornea, the other is the retina. And with good reason, because if blood vessels do invade these sites, they can cause blindness. But how is this achieved? It turns out that both sites produce a soluble molecule that soaks up - and therefore blocks - the signal that would normally trigger blood vessels to form. Chris Smith spoke to Bala Ambati from the University of Utah...
discouraged from growing: one is the cornea, the other is the retina. And with good reason, because if blood vessels do invade these sites, they can cause blindness. But how is this achieved? It turns out that both sites produce a soluble molecule that soaks up - and therefore blocks - the signal that would normally trigger blood vessels to form. Chris Smith spoke to Bala Ambati from the University of Utah...
Bala - When you look at the eye, it has several important yet, almost contradictory missions. The cornea in the front of the eye has to stay clear, it has to stay strong, it has to focus and it has to do all that without having any blood vessels that can give it nutrition or oxygen. Now in the retina, at the very back of the eye is what's called the photoreceptor layer and the photoreceptors are the rods and cones that convert light into sight. Those rods and cones use a lot of oxygen. They're actually the most metabolically active cells in the body. And so, they are situated right next to the choroid - the blood vessel tissue network that has the highest blood flow of any vascular network in the body. And so, you have these photoreceptors that demand high oxygen, situated right next to this incredibly rich blood vessel network. Yet, that layer of the retina should not have blood vessels because as soon as blood vessels come in, that can distort the vision. And so, you have these contradictory imperatives - clarity versus oxygen demand.
Chris - How did you approach that and begin to ask, what keeps the blood vessels out of the retina, despite its very high oxygen demand. and equally the cornea?
Bala - I started looking at the cornea and realised that the cornea expresses several different VEGF receptors and VEGF is a vascular endothelial growth factor, but interestingly enough, it expressed one particular form of VEGF receptor called sFlt-1, soluble VEGF receptor-1. We looked at multiple mammals. One of the very few mammals that in nature, does not have a clear cornea is the Florida manatee and it actually has a vascularised cornea. A couple of mouse strains that are genetically mutated also have vascularised cornea. And so, we found that in these mutant mice and in the manatee, this sFlt-1 was missing. And we also found that restoring sFlt-1 to mice that were lacking in the molecule helped restore corneal clarity and the lack of blood vessels.
Chris - Is the model of a hypothesis then that the cornea in this instance is secreting this molecule which has the effect of soaking up a factor which would otherwise trigger the growth of blood vessels? It's locking it away, so that the blood vessels do not grow.
Bala - Precisely.
Chris - And so, then I guess, you must've thought, well, if that's what's going on in the front of the eye, could the same trick be playing out in the back of the eye, in the retina? So, is there a soluble form of this - for want of a better phrase - blotting paper for VEGF, the factor that makes blood vessels in the back of the eye, preventing it from acting there in the retina too?
Bala - Absolutely. So, the soluble VEGF receptor is a decoy - or as you put it, blotting paper - so it serves as a sink for VEGF. And so, our very first preliminary experiment was just to see, well is this molecule expressed in the retina? And interestingly enough, it was expressed in the retinal pigment epithelium which is the layer that is underneath the rods and cones, and just above the choroid. And so, that thin red line if you will, of RPE tissue is what keeps those choroidal blood vessels out of the retina and the RPE does indeed express sFlt at high levels.
Chris - So, I suppose it could be regarded as the million dollar question - if you look in people who've got disease in their retina, diseases characterised by new blood vessels growing in - things like wet macular degeneration, do they lose the secretion of this VEGF blotting paper, the sFlt signal which allows those blood vessels to invade in the way they do pathologically?
Bala - Exactly. I'd say, that's not just a one million dollar question, but it affects 10 million Americans and 2 million Britons. And so, we've proceeded to examine what happens in patients with disease then we did a number of experiments in mice, and we found that knocking down this molecule sFlt, allows these blood vessels, the choroidal blood vessels to invade the retina.
Chris - Do you think that if we were to go in and therefore turn that signal on more or enhance it in people with disease, we could arrest the disease process and do it in a safe way?
Bala - Correct. There's a sister paper that came out in ACS Nano, just a couple of months ago where we present the data on that exact same question. We found that in both a mouse model of macular degeneration and a monkey model of macular degeneration, that if we delivered nano particles loaded with a subunit of Flt-1, inject it intravenously, not into the eye but just their systemic vein, it could cause regression of these choroid blood vessels growing into the retina in both mice and monkeys.

22:22 - Watching T cell Receptors at Work
Watching T cell Receptors at Work
with Geoff O'Donaghue
What do you do if you want to see how an immune T-cell responds to meeting the antigen molecules that it's programmed to detect?
Geoff - My name is Geoff O'donoghue. I'm a post doc at UC Berkeley in the lab of Jay Groves. The general question we were interested by was how the immune system is able to detect form pathogens, things like virus and bacteria, and how this occurs at the molecular level. So, we were looking at T-cells, CD4 or helper T-cells and we were looking at the T-cell receptor and the T-cell receptor binds a pathogenic molecule on the surface of another cell. It's just like a lock and a key basically at the receptor on the T-cell surface is a receptor and it has a ligand that it's looking for. Once it finds that, it binds and initiate an immune respon se specifically only for that one pathogen.
se specifically only for that one pathogen.
Chris - So, you look at these T-cells, CD4 cells specifically. How do you study that interaction?
Geoff - We study the interaction using optical imaging. Ideally, we would look at the interaction at the interphase between a living T-cell and what's called an antigen presenting cell. But that's a very difficult thing to image because you have two curved surfaces trying to image the interphase between those two curved surfaces with high temporal and spatial precision is very difficult. The trick that we use is to replace the antigen presenting cell with, basically, a biomimetic surface that is flat. We put on the surface the support lipid bilayer. So, this is a very good mimic of a cell. Attached to this support lipid bilayer, we attached the pathogenic molecules. In those molecules are free to diffuse laterally in two dimensions and then interact the T-cells once we flowed those on top of this surface.
Chris - And you can see where these pathogenic molecules are going, can you?
Geoff - Yes. What we did was label each molecule on the surface with a tracer molecule where we can relatively easily observe. We basically observed a field of these individual molecules, diffusing around in two dimensions. Once we flowed the T-cells in, the T-cells bind some of those pathogenic molecules on a surface. Once that occurs, we observed that instead of diffusing around, the tracer molecules on the surface flow way down. And then we can relatively easily observe. The way that technique that we use was basically to use a very long camera exposure time, the images are similar to the long time lapse images you see of a city where the cars on a road are blurred out because they're moving so quickly, but the buildings in the background are very stationary and very clearly resolved. We took the same approach to distinguish between single molecules that are bound to their ligands in the living T-cell and once they were unbound, instantly diffusing on the surface.
Chris - And so, this enables you to tell when a T-cell has interacted with one of those ligand molecules on the biomimetic surface. Are you then able to tell how the T-cell responds to that interaction?
Geoff - Yes. So, the first thing we do is measure the number of seconds that we observe this interaction happening. That tells us something about the time component that the T-cell is detecting. And then we're looking at a molecule inside the cell which is an enzyme and induces a cascade of chemical reactions inside the T-cell. And so, what we did was two-colour single molecule imaging. We looked at the pathogen on the surface and the catalyst inside the cell, and we measured the number of molecules that are recruited to the reaction site involved.
Chris - So, you can very neatly here, identify when the T-cell docks or binds onto the thing on the cell surface, or the pretend cell surface. You can see exactly when the T-cell begins to respond and how vigorously a T-cell responds. That's amazing! So, what did you find?
Geoff - What emerged was that interactions between the pathogenic material and its receptor were surprisingly long lived. It wasn't clear from the previous literature whether or not these would be short much, much less than a second or seconds to minutes long. We discovered that these interactions occurred for a surprisingly long time. what we also observed is that one molecule could induce a chemical cascade inside the T-cell. The T-cell is very sensitive and can detect single molecules or a handful of molecules that will induce a chemical reaction inside the cell. The next question is to look at not just one pathogen but to see how T-cells respond to a panel of these pathogens and then kind of get at that question, is a large number of short interactions equivalent to a small number of very long lived interactions. That's I think the next step and that would go some way towards kind of really getting at the question of how T-cells distinguish between self and non-self.

27:52 - Inside eLife, July 2013
Inside eLife, July 2013
with Peter Rodgers, eLife Features Editor
 Features editor Peter Rogers has a selection of some choice morsels he discusses with Chris, that have appeared this month in the journal.
Features editor Peter Rogers has a selection of some choice morsels he discusses with Chris, that have appeared this month in the journal.
Peter - Okay, well I'm going to talk about three papers. The first is actually a point of view article by Ian Boyd who is the Chief Scientific to DEFRA, The Department of Environment, Food, and Rural Affairs in the UK. Now, Ian Boyd is very busy at the moment with issues related to bovine TB. But the article he's written for eLife is a more general piece about how scientists interact with government and are involved in the making of government policy. It's called 'Making Science Count in Government'.
Chris - What points does he make?
Peter - I have described it to people as tough love and it doesn't pull his punches when he tells scientists what they need to do. I'm just going to read directly from the article. He says, "His experience suggests to me that there can be confusion amongst some scientists about their role and also about how they can bring their influence to bear most effectively." And the thing he stresses is that, science is just one of many inputs that have to be taken into account on government sent policy and they're sort of social and electoral, ethical, cultural, legal, all sorts of other issues which need to be taken into account. So that it might seem to scientists that their concern aren't foremost in decision - which is a reality because there are other things that need to be taken into account as well. It gives an example of the complexity and the difficulty of the whole area in bovine TB where he says, "within the UK and Ireland, there are actually 4 different policies being followed to treat the problem, all presumably based on the same scientific evidence". So, in England, it's proactive badger culling and Wales, it's vaccination, in Northern Ireland it's test and then vaccination, and then in the Republic of Ireland, it's reactive badger culling. There's not one single correct solution to many of these problems and they are quite complex.
Chris - How has that gone down with the scientists?
Peter - Well, we've just published the article a couple of days ago, so we're still waiting for the reaction.
Chris - How do you expect it to go down with the scientists?
Peter - Well, I think it will come as a surprise to them in the first instance, but he speaks very clearly and very plainly. He is at the heart of government involved in the setting of these policies. So, I think they'd be well-advised to listen and pay attention to what he has to say.
Chris - A lot of scientists think that government doesn't understand science and they think that they're not very well listened to. So, is he saying the scientists need to be more vocal and be more forthright or is he just saying, "Just accept the fact that your voice is one of many. Don't stop saying what you're saying, but don't expect always to be heard."
Peter - I think he's saying that there is sort of an apparatus and a mechanism of advisory committees and chief scientific advisers like he himself and that scientists should go through these bodies. They should recognise that the decisions won't always be what the scientists may want them to have been and that if they complain too much when that happens, it risks being counterproductive and maybe the scientists won't be asked for their advice in the future.
Chris - I shall have to check out the article in my own mind up which is very important science. Now, moving on to look at some of the other research that's been published this month, you have an item on ammonium sensing.
Peter - That's right, yeah. So, ammonium is the ion of ammonia. So, the chemical formula is NH4 and it's positively charged. It's a very important source of nitrogen. Ammonium can be moved in and out of cells either by transport proteins or through ion channels that move lots of other ions and water molecules in and out of cells. So, to actually get the grips with the levels of ammonium, Wolf Frommer and his colleagues at the Carnegie Institute of Science developed a sensor based on green fluorescent protein which they introduced into one of these transport proteins that moves the ammonium within the cells. The level of intensity coming out from this protein sensor basically changed in response to the levels of ammonium.
Chris - Is this in eukaryotic cells or prokaryotic cells - bacteria?
Peter - They did it actually in a variety of cells. They did it in frog cells and yeast cells. One of the other things that's interesting about this is that some of the mechanisms for the transport of ammonium to be conserved throughout many different types of cells. And the hope is that this technique will be able to use wide variety of species.
Chris - And talking about conservation of things through evolutionary time, you also got some stuff on glucose and how cells use that.
Peter - That's right. So, Dawn Thompson and our colleagues at the Broad Institute have been studying gene regulation which is a process by which genes are switched on and off in cells. What they did is they measured profiles of messenger RNA and 15 different species of yeast evolve from a single ancestor over the past 300 billion years. So, the patterns of gene regulation have sort of diverged over time as you might expect, but the change particularly following either changes in the lifestyle of the yeast are events called whole genome duplication events which as the name suggest, during meiosis, the whole genome is just duplicated. They find those two things are the biggest influence on the gene regulation. And there's an interesting connection which they sort of elude to and it needs further work. Most cells use glucose as a source of energy that doesn't lead to the cell proliferating and dividing. But some of these cells use glucose to divide and proliferate which is very like what happens in cancer. So that there were interesting parallels which could be explored and might tell us something more about cancer as well.
- Previous Man-powered Helicopter
- Next Let's talk about sex









Comments
Add a comment