The Science of the Seriously Small
This week, we're studying the science of the seriously small - nanotechnology. We'll find out how tiny, flexible electronics could be implanted under the skin to restore lost sensation, and how tiny protein covered silicon "diving boards" can show us how superbugs evade antibiotics. Also, how sheets of carbon just one atom thick can be used to read the entire human genome in just a couple of hours, and how nanotech "motherships" can deliver exactly the right amount of drug, directly to where it's needed. Plus, the plant genome that could solve the food crisis, how our fingerprints help us to feel fine textures, and how a new way to make LEDs could slash our household bills. And, as if that wasn't enough, in Kitchen Science Dave will be looking for silver in soot!
In this episode

Pointing out how fingerprints help us feel fine details
In a piece of true science detective work, researchers at the Laboratoire de Physique Statistique in Paris have found another reason why we have fingerprints.
 It's been known for a while that the distinctive ridges on the pads of our fingers help us to grip things, but now Julien Schiebert and colleagues have shown that fingerprints also help us to feel fine textures and tiny objects, less that 200 micrometers, through vibrations.
It's been known for a while that the distinctive ridges on the pads of our fingers help us to grip things, but now Julien Schiebert and colleagues have shown that fingerprints also help us to feel fine textures and tiny objects, less that 200 micrometers, through vibrations.
Writing in the journal Science, they developed a special mechanical sensor which is fitted with a rubbery cap to act like a fingertip. Using either smooth caps, or ones ridged to simulate fingerprints, they rubbed their artificial finger across finely textured surfaces.
They found that the cap with a 'fingerprint' was made to vibrate by the contact, at a frequency of around 250Hz, that's 250 vibrations per second. This coincides with the sensitive range of a type of nerve endings found in the skin, called "Pacinian corpuscles".
Two types of nerve ending are known to be involved in detecting texture, slow reacting nerves are responsible for identifying relatively coarse textures by detecting different pressures at different places on the skin, while fine details are reported by the Pacinian nerve endings.
They have only tested this with a series of straight, parallel ridges, so not exactly the same as the swirly lines of our fingerprints, but their findings suggest that our fingerprints actually fine tune the vibrations, selectively amplifying certain frequencies to ensure our nerves can pick up the fine details.
Writing the world’s smallest letters
Keeping with our theme of nanotechnology this week, researchers at Stanford University in the States have managed to write the smallest letters ever - assembled from subatomic particles just 0.3 nanometres in size. The researchers are particularly pleased with their achievement, because it was Stanford scientists that first created the world's smallest writing in 1985, but lost the record in 1990 to IBM, when they famously arranged xenon atoms to spell out the company's name. Now they have it back.
But how did they do it? The researchers encoded the letters "S" and "U" (standing for Stanford University) by using the interference pattern of electron waves on the surface of a film of copper. The technique actually projects a tiny hologram of the letters, which can only be seen using a very powerful microscope.
Writing about their work in the journal Nature Nanotechnology, the Stanford team's letters are more than four times smaller than the IBM initials, and were created using a scanning tunnelling microscope, which can be used to push atoms around. The scientists used it to put individual molecules of carbon monoxide in a special pattern on a film of copper the size of a fingernail.
Electrons are constantly whizzing around on the copper, because it is a metal. And because electrons can act as waves, as well as the more traditional view that they are particles, the electron waves get shaped by the carbon monoxide, and interfere with each other like ripples in a pond. These interference patterns depend on the position of the carbon monoxide molecules on the copper surface. So eventually they create a consistent pattern that can be read, like a molecular hologram.
It all sounds quite nerdy, but the technique could be very important for the future of computing, as it would enable information to be stored at a very high density in small chips. For example, the researchers could create different holograms on the same chip by using different electron wavelengths, increasing the amount of information that can be stored, and pushing it beyond current boundaries.

Mob rule to scare away cuckoos
Researchers studying Reed warblers have found out that mob rule can avoid being cuckolded by cuckoos.
 Cuckoos live a parasitic lifestyle - laying eggs in the nest of other birds and letting them spend their time and resources bringing up their young. From an evolutionary perspective, it's a good trick if you can get away with it, but if you're the victim you're wasting your own resources on someone else's DNA.
Cuckoos live a parasitic lifestyle - laying eggs in the nest of other birds and letting them spend their time and resources bringing up their young. From an evolutionary perspective, it's a good trick if you can get away with it, but if you're the victim you're wasting your own resources on someone else's DNA.
Writing in Current Biology, Cambridge University researcher Nick Davies reports on how Reed Warblers use mobbing techniques to keep the parasitic cuckoos away from their nests.
Mobbing is a risky strategy - it's energy intensive and it exposes you to predators, and may not prevent the cuckoo from getting through. Worse still, sometimes they mistakenly mob a sparrowhawk, which looks a bit like a cuckoo and actually prays on reed warblers.
Some birds would save their energy, and just reject any eggs that don't look like their own, but the cuckoos have evolved to be able to lay 'mimic' eggs which look similar, establishing an evolutionary arms race between parasite and host.
By placing model cuckoos near the reed warbler's egg-bearing nests, Davies and colleagues could observe how the warblers attempted to defend their nests. About half the time, the warblers became aggressive and attempted to mob the cuckoos. In the high risk areas, this made them far less likely to be subject to a cuckoo visit than their more peaceful neighbours.
Significantly, in areas where the Warblers were at much lower risk of being parasitized, they were far less likely to show this mobbing behavior - in fact, in those areas mobbing was likely to attract cuckoos, rather than scare them away
Reed warblers also reserved mobbing behavior only for cuckoos, showing that they adapt their nest defense strategy according to their conditions, not unlike our own military!

Turning up the heat on cereal genome
It's clear that the global climate is changing, and this is having a big impact on food supplies. For example, if the climate changes in a major crop-growing region, it may not be possible to grow that crop successfully any more. So scientists are investigating whether people living in dry regions - that are only getting drier - can grow alternatives to wheat and other food crops.
 One such alternative is a plant called sorghum. This is a type of grass that originally came from Africa, and it grows well under hot and dry conditions. Now farmers in warm parts of America, Asia and Europe are growing sorghum for food and animal fodder, as well as for using in biofuels. Not only that, but it can be burnt to provide energy.
One such alternative is a plant called sorghum. This is a type of grass that originally came from Africa, and it grows well under hot and dry conditions. Now farmers in warm parts of America, Asia and Europe are growing sorghum for food and animal fodder, as well as for using in biofuels. Not only that, but it can be burnt to provide energy.
It sounds like an all-round wonder-plant, and in order to uncover the secrets to its versatility and hardiness, researchers in Munich have analysed the whole sorghum genome. This is the first time the genome of a plant of African origin has been sequenced.
Publishing their results in the latest issue of the journal Nature, the scientists say that their results will help us understand more about how plants like sorghum resist drought and high temperatures, and could help with the development of hardier versions of other crops in the future.
And the new data will also enable researchers to compare the genome of sorghum with rice and maize, two important crop plants that have had their genomes sequenced. This will tell us a lot about how crop plants evolve, and the genes that give them their specific properties.

09:35 - New LEDs to Slash household bills
New LEDs to Slash household bills
with Professor Colin Humphreys, University of Cambridge
Ben - There's a new way to make LEDs and this could slash household lighting bills and help to make clean drinking water accessible to everybody. Professor Colin Humphreys from the University of Cambridge joins us now on the line. Hi, Colin.
Colin - Hi.
Ben - Tell us a bit about this. We've had LEDs for a long time. They are already turning up in torches, in home lighting. What's the new method that you've got?
 Colin - They've been around for some time. They're not really in home and office lighting and the reason is they're too expensive. All the LEDs you can buy in the shops now are grown on a sapphire substrate and sapphire is quite expensive. What we've done is to develop a method for growing these LEDs on a silicon substrate. In fact we're growing them on a six inch substrate wafer instead of on a two inch sapphire wafer. That's going to be bringing costs down by a factor of ten or so, a really big reduction.
Colin - They've been around for some time. They're not really in home and office lighting and the reason is they're too expensive. All the LEDs you can buy in the shops now are grown on a sapphire substrate and sapphire is quite expensive. What we've done is to develop a method for growing these LEDs on a silicon substrate. In fact we're growing them on a six inch substrate wafer instead of on a two inch sapphire wafer. That's going to be bringing costs down by a factor of ten or so, a really big reduction.
Ben - Wow. These LEDs are very energy efficient, I understand. How will it compare to a normal incandescent light bulb?
Colin - They're very energy efficient and they're really going to help global warming. In fact, they'll help it much more than wind power will. In terms of an incandescent light bulb we're aiming to be twelve times as efficient as a tungsten light bulb and we're aiming to be 3 times as efficient as a low energy light bulb. Already they're more efficient than a low energy light bulb.
Ben - Is that for the same brightness as well?
Colin - That's for the same brightness, absolutely. We want to make them better quality and better bright light than the low energy bright light ones, which as you know are not very popular. They're a popular cause for divorce in the country now, I'm told, low energy light bulbs!
Ben - Hopefully your gallium nitride LED light won't be a cause for divorce but I've also heard these could be used to make clean drinking water. I understand what you can do with these is make ultraviolet light.
Colin - That's right so the light emission from the gallium nitride - we actually add some indium to it to get visible light emission. If you add aluminium you can get deep ultraviolet. Deep ultraviolet has certain wavelengths, it's about 270nm. It destroys the nucleic acid in both DNA and RNA and it stops viruses and bacteria from reproducing so it effectively kills them. If we can make LEDs that emit this deep UV we can kill all known viruses, all known bacteria and you could put a ring of these LEDs on the inside of a water pipe coming into a home in the third world. Water riddled with bacteria and viruses, you can make it harmless. Also it could be useful for our country as well, particularly for third world people.
Ben - And we could use them in hospitals as well to ensure things are sterile without having to go through the chemical cleansing that we do now. As they are so efficient does this mean we can set this up with a solar panel and make this water purification very portable?
Colin - Absolutely. That's absolutely right. These are very efficient. They'll run of 4 volts, which is ideal for a solar panel, you have a solar panel and a battery connected as well if you like and then have these connected to that. You have these for lighting in the developing world but also for water purification in the developing world.
Ben - These sound fantastic but what's different about gallium nitride that allows us to make this seemingly wider range of frequencies? If we can make UV that we couldn't before. Why is gallium nitride so special?
Colin - Gallium nitride's called a wideband gap semiconductor. Before it came along the only light emission which you get from semiconductors was in the infrared and in the red and rather weakly in the green and the yellow. Silicon doesn't emit light anyway. Gallium arsenide emits light and indium phosphate but they're narrow band gap materials. If it's a much wider band gap material, gallium nitride itself emits near ultraviolet and then there's another material called indium nitride which emits in the infrared. If you mix those two together you can get any energy you want from the near-infrared going right through the visible spectrum to near-ultraviolet. If you add some aluminium to it you can go really into deep ultraviolet. This is a new material system. It's man-made. It can cover this range of the electro-magnetic spectrum that we've never had before form a solid state semiconductor.
Ben - This sounds quite incredible. When should we expect to see these on the market?
Colin - For the home and office lighting -scientists always predict thing will happen before they're going to happen! I think within the next five years, certainly. Maybe two or three years. The UV problem is more difficult to solve. We've already got the right wavelength being emitted but the intensity at the moment is too low. We've got to push up that intensity. I think realistically that may be 5-10 years. I really believe it's going to happen.
Ben - Clearly getting clean and drinkable water to everyone in the world is going to be a challenge and I suspect unfortunately it will still be a challenge in those 5-10 years. Good luck, I hope we get them to market as soon as possible!

27:04 - Nano Diving Boards for Bacteria
Nano Diving Boards for Bacteria
with Dr Rachel McKendry, London Centre for Nanotechnology
Meera - This week I'm at the London Centre for Nanotechnology which is a joint venture of University College London and imperial College London. I'm here to see a new way of screening antibiotics which could help speed up the search for antibiotics against the ever-increasing hospital superbugs like MRSA. I'm here with Rachel McKendry, a reader in biomechanical nanoscience here at the London centre for nanotechnology and she helped developed this technique. Rachel, how have you been screening the effect of antibiotics against bacteria?
Rachel - Well, we've been developing a novel nanomechanical approach to study antibiotics and to understand more about their modes of action and the mechanisms of superbug resistance.
 Meera - And how have you looked into this?
Meera - And how have you looked into this?
Rachel - We use arrays of tiny silicon diving boards called cantilevers which we coat with different peptides found in bacterial cell walls. We then inject different antibiotics in solution and for our studies we focussed on vancomycin which is, in fact, one of the most powerful antibiotics that we have in the battle against resistant superbugs.
Meera - How does putting bacterial proteins on a diving board and then putting a solution of antibiotics help you learn about the effect of these antibiotics on the proteins?
Rachel - When the antibiotic binds to the peptide on the cantilever it causes the cantilever to bend by a very tiny amount, just a few nanometres. But we can detect this by shining a light on the very end of the cantilever and measuring its position on a photosensitive detector. What we've found is the amount of bending is proportional to the concentration of antibiotic in solution. From this we can learn about the strength of the interaction, the binding constant which is a measure of how, essentially how powerful the drug is in the body.
Meera - Does this mean that the greater amount of bending you measure the greater the damage to the bacteria?
Rachel - Yes, exactly. That's our concept, that the biding generates huge mechanical consequences on the bacteria.
Meera - And just here in front of us we've got an example of one of the silicon chips that you used to mount the bacterial proteins onto. It's tiny but it's about half a centimetre long so we can actually see it.
Rachel - You're right in the sense. These are relatively large objects. They can be seen with the eye but if you look very closely at the top end you can see the silicon cantilever arrays. These are the diving boards at the end. It's their thickness that's the critical factor in determining their properties. They're 500microns long. That's half a millimetre long, 100 microns wide - that's typically the width of a human hair but the thickness is only 900nanometres. This means that it can detect very tiny changes in forces at the surface of the cantilever.
Meera - A key part of this experiment was that you used bacterial protein from bacteria that were resistant to antibiotics and also ones that were sensitive so you could see the difference between resistant and sensitive bacterial strains.
Rachel - Yes, the peptides differ by a single amino acid mutation and the mutation confers a deceptively simple change in the way that the drug works. It deletes a single hydrogen bond from the pocket between the antibiotic and the peptide and the binding of the antibiotic to the peptide found in drug-resistant bacteria is 1000-fold weaker than those found in drug-sensitive bacteria. This renders the drug therapeutically useless so we've been fascinated with understanding this process and essentially hope in the future that we can design new antibiotics that combine to these peptides found in resistant superbugs.
 Meera - One of the true benefits of this particular technique is that it can screen the effectiveness of antibiotics quite quickly. What makes this so much quicker than the other technique currently being used?
Meera - One of the true benefits of this particular technique is that it can screen the effectiveness of antibiotics quite quickly. What makes this so much quicker than the other technique currently being used?
Rachel - Firstly that it's label-free. This means that you don't need to use fluorescent or radioactive probes as you might have to with other technologies and this has an advantage in terms of time and cost. But also labels can potentially perturb the way a biomolecule works. Other advantages are that they are immediately compatible with silicon microfabrication technology and what I mean by that is that it's possible to scale-up the number of cantilevers, readily for high density arrays to screen potentially thousands of drugs per hour. We hope that it will potentially provide a new way to understand how antibiotics work and hopefully develop more antibiotics in the future.

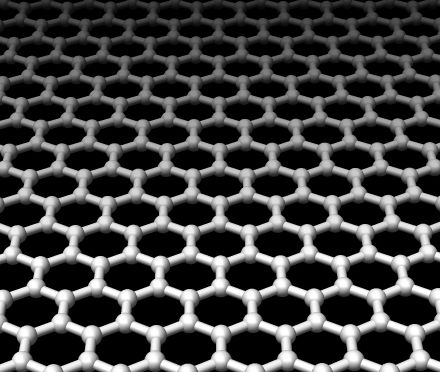
32:41 - Rapid DNA Reading with Graphene
Rapid DNA Reading with Graphene
with Dr Henk Postma, CSU Northridge
Ben - As an understanding of our genes and that of our food and everything else becomes more important in knowing what diseases we may develop and, in fact, how to treat them and avoid them - fast an efficient ways to sequence our genome becomes ever more important. Dr Henk Postma for the California State University at Northridge thinks that nanotechnology could have the answer. Hello Henke, thanks for joining us. How would we use nanotechnology to read DNA?
 Henk - The idea has been kicked around by some scientists for a few years and the idea is that if you would have somehow separate electrodes connected to a single base of DNA molecules then if you would read its conductance then you could tell the difference between the types of A-C-G-T in a chain. If you could somehow scan your electrodes on the molecule you could read off the sequence. What we are planning to do is we are planning to use electrodes that are fixed and then are immersed in a reservoir of liquid. We would introduce DNA on one side then we would apply an electric field and the electric field would drive the DNA molecules through that pair of electrodes. While it goes through you read off the conductance.
Henk - The idea has been kicked around by some scientists for a few years and the idea is that if you would have somehow separate electrodes connected to a single base of DNA molecules then if you would read its conductance then you could tell the difference between the types of A-C-G-T in a chain. If you could somehow scan your electrodes on the molecule you could read off the sequence. What we are planning to do is we are planning to use electrodes that are fixed and then are immersed in a reservoir of liquid. We would introduce DNA on one side then we would apply an electric field and the electric field would drive the DNA molecules through that pair of electrodes. While it goes through you read off the conductance.
It's deceptively simple but it turns out it's actually quite hard to do and people have been trying to do this with big electrodes. If you make electrodes out of gold materials they typically are very thick, let's say about 20nm thick. Then your DNA molecule has a distance between the different bases of 0.3nm. You would have at any point in time 60 bases in between your electrodes so it's very hard to resolve any kind of electrical signal due to a single base.
Ben - So you'd get an indication of what bases might be between your electrodes but you wouldn't be able to read each one?
Henk - Exactly. You'd get some kind of statistical measure of the average, perhaps. What I've been proposing is to use graphene as the electrode. Graphene is a new material that was discovered by Andre Geim in Manchester, in your country, about 5 years ago and it's a single layer of carbon. It is extremely robust, it is a very good electrical conductor and because it's a single atom thick if you make electrodes out of those you would be able to resolve the single bases inside the DNA molecule. That's the basic idea of what I'm working on.
Ben - It sounds like a deceptively simple idea but you also imply that it's just an idea at the moment. What do you think the challenges will actually be when you can make this happen?
Henk - A few of the things have been solved already by several people in the world. One of the things you need to do is you need to be able to make a membrane that can be immersed in a liquid and survive the typical capillary action that is associated with having something as thin as that in the liquid. People have solved that. They've made thin membranes. One of the things you need to do is you need to make a tiny gap and we've identified a few potential technologies that allow us to make a thin gap. That's the first thing that we're actually working on right now. That's the first thing that we need to be able to do.
 Ben - This will need to be a gap that's exactly the right size to let DNA through, one base at a time.
Ben - This will need to be a gap that's exactly the right size to let DNA through, one base at a time.
Henk - Exactly. It has to line up very nicely because if you would even make you gap about one nanometre thicker, wider than it is in the ideal case your current drops by several orders of magnitude. It would be very hard to detect any kind of signal. That would be the first challenge, I would say.
Ben - What are the advantages of doing this? How quickly could you actually sequence the human genome? We know there are 24,000-ish genes that actually work and a load of stuff that may or may not be junk. How long do you think it would take?
Henk - Well, actually the junk DNA is actually a very interesting issue. We're proposing to sequence the whole thing regardless of whether we consider something junk or not. That is an open question, whether junk DNA ha'ssome kind of a function. Extrapolating from our read-time for a single base I estimate it will be about 3 micro seconds so, if you would be able to use such a device for one single DNA molecule of the human, it would take about 2 hours to read the whole thing.
Ben - That's really incredible. Just one last question, how much is it likely to cost to sequence one human genome?
Henke - Well you would have to get some electrical equipment, make some material. Graphene actually is surprisingly cheap. I'm not sure, my lab is very expensive but I presume that if you want to do it again you don't want to make any extra investments. I won't be able to put a number on it! That's something that has to be figured out later but because it's a single device and there's no extra preparation that is needed for the DNA material you can just put it in the liquid and you'll be done with it. You don't need to do any PCR amplification or radio labelling or fluorescent read-out or any gel electrophoresis. All those techniques are not required. It's a very basic experiment, actually.
39:06 - Mothership for Nanotechnology
Mothership for Nanotechnology
with Professor Michael Sailor, UCSD
Ben - I caught up with Professor Michael Sailor from the University of California at San Diego, another person enjoying the sun while we're in the snow, to find out how you can convince such tiny particles to be your drug deliverers.
 Michael - To get a material to deliver a drug one of the themes that we've been developing is so-called mothership for nanotechnology. We build a nanostructure that's fairly large. It's still a nanostructure but it's large enough to hold a smaller component inside it. What will make these porous silicon-based nanostructures that can contain a drug. One of the really favourite projects right now, a really exciting one that I have going on is working with a guy named bill Freeman who's at the Jacob Retina Centre here at UC San Diego. He sees patients every day with a disease called macular degeneration. That's a disease of the eye, typically age-related. One of the main causes of blindness in older folks. There are no drugs available that you can give to a patient as a pill and have it get through the stomach and make it to the eye. So they unfortunately have to take their patients and stick a needle in their eye with the drug. The problem with that is there's a chance of infection and once you put the drug in the eye typically these kinds of drugs will only last for a month or two before they're all gone and then the patient has to come back in and be injected again. Bill approached me with this problem and he said the only way we can keep the drug in the patient's eye for a therapeutically useful time is to massively overdose them. We have to inject a lot of drug in the eye. If we put a high dose in the patient's very susceptible to problems. There's probability of strokes and other complications that come from having too high a concentration of drug. He knew we were working on these little nanoparticles that had pores in them. He said can you put the drug inside those structures, lock it up in there, keep it away from the body, sequester it until it's needed and then have it leach out slowly? So the level of drug concentration in the body is much lower - still above the therapeutic level so it's treating the disease but not so high that it does any damage. What we are now developing are materials that can stay in the eye - at least in the rabbit eye for eight months.
Michael - To get a material to deliver a drug one of the themes that we've been developing is so-called mothership for nanotechnology. We build a nanostructure that's fairly large. It's still a nanostructure but it's large enough to hold a smaller component inside it. What will make these porous silicon-based nanostructures that can contain a drug. One of the really favourite projects right now, a really exciting one that I have going on is working with a guy named bill Freeman who's at the Jacob Retina Centre here at UC San Diego. He sees patients every day with a disease called macular degeneration. That's a disease of the eye, typically age-related. One of the main causes of blindness in older folks. There are no drugs available that you can give to a patient as a pill and have it get through the stomach and make it to the eye. So they unfortunately have to take their patients and stick a needle in their eye with the drug. The problem with that is there's a chance of infection and once you put the drug in the eye typically these kinds of drugs will only last for a month or two before they're all gone and then the patient has to come back in and be injected again. Bill approached me with this problem and he said the only way we can keep the drug in the patient's eye for a therapeutically useful time is to massively overdose them. We have to inject a lot of drug in the eye. If we put a high dose in the patient's very susceptible to problems. There's probability of strokes and other complications that come from having too high a concentration of drug. He knew we were working on these little nanoparticles that had pores in them. He said can you put the drug inside those structures, lock it up in there, keep it away from the body, sequester it until it's needed and then have it leach out slowly? So the level of drug concentration in the body is much lower - still above the therapeutic level so it's treating the disease but not so high that it does any damage. What we are now developing are materials that can stay in the eye - at least in the rabbit eye for eight months.
Ben - How does it avoid the processes that usually recycle the fluid in the eye and would usually take away the drug that they'd put in there.
Michael - If you had a material that you're putting into the eye and it stayed there even after its effective time was done after it's already delivered its drug then it eventually ends up you're going to start occluding the vision. A real key thing for that was to have these materials definitely dissolve away into nothing. It has to dissolve away slow enough that the drug locked inside is delivered in a fairly smooth and slow fashion.
Ben - This should mean that people suffering from AMD would at least have one nanoparticle administered every six months rather than an injection of an excess of drug every month.
Michael - Well, we still have to put a lot of drug in the eye so it wouldn't be one nanoparticle. We would be injecting large quantities of nanoparticles: roughly the same volume that the doctor now injects into a patient. They key thing is that when that drug goes in to the 100microlitre injection that's going into the eye it's not all instantly available. Some of it is locked away and it's releasing over a much slower time period.
 Ben - When do you expect that the people will be queuing up for their clinics to have their nanoparticle injections?
Ben - When do you expect that the people will be queuing up for their clinics to have their nanoparticle injections?
Michael - That's a really tough question. We would like to get into the clinic within a year. We're at the stage right now where we're doing experiments with rabbits. As you know with any material; nanomatrials in particular we want to be very careful we're not doing more harm than good. The problems that may come up, infections, longer-term issues - do materials really degrade away to nothing, are they getting eliminated completely? There's a lot of work that still needs to be done. One of the real challenges here with the nanomaterial is even if you think it's harmless and I'm sitting here saying these are made of silicon dioxide or iron oxide and that's a material the body can accept and degrade smoothly there are great examples of nanomaterials that are made of things that are completely harmless that are really nasty. For instance, carbon nanotubes do not degrade in the body at all. Unless your body can eliminate them somehow if it gets stuck somewhere it's there forever. If you look at asbestos, the chemical composition of it is quite harmless itself: aluminium, oxygen and some iron initially so kind of the same sort of constituents that we're talking about here. Why is that particular alumino-silicate, that asbestos mineral so harmful? Because it's in a nanostructure that doesn't degrade it's actually quite harmful. Even though its elemental components are not toxic when that material gets into the body and lungs in particular for asbestos it sits there and doesn't go away. The body can't dissolve it and so those fibres will stay lodged in the lungs, make lesions and cause problems. That's a real moral to the story of nanomaterials. It's not good enough to just say the elements are harmless. You've got to know that the material itself will go away.

Spit and Polish?
We put this to Chris Powley-Williams, Leatherwise in Northampton, UK...
We're quite intrigued by the question posed by Dave as to why spit and polish is so successful on your shoes. The reason is all to do with protein.
Traditionally in the leather industry, when we wanted a very highly glazed or highly polished shoe we used to use the protein that you find in milk, and that's casein.
It would have been applied to the leather surface and it would have been fixed in place with formaldehyde or formalin, some people would know that as. Then we would have used a glass block and glazed the actual finish and that would have given you this high polish effect.
In effect that's what Dave is doing every time he spits on his shoes. In your saliva there's a high amount of protein. By spitting on your shoes and then rubbing it up to a polish you're actually trying to glaze the protein in the same way we would have done traditionally, many years ago.
He then poses the question, well nowadays how are finishes different? We tend not to use the protein finishes. They're very brittle. We're not allowed to use formaldehyde in the same way we used to use it, and so we tend to use a lot of polymer finishes and nitrocellulose, polyurethane, things like that. We can achieve the same levels of gloss but in quite a different way.
One interesting fact we came across ourselves was that, actually, it's best to clean your shoes when you're happy! The reason being that the protein in your spit or saliva actually decreases if you're feeling depressed and increases when you're feeling happy. If you want a really good gloss on your shoes only spit on them when you're in a good mood. That should solve all your problems!





Comments
Add a comment