Why can some people stay fit as a fiddle, while other seem to catch every bug that's going? As you might expect, the answer lies in our genes. Plus, a multitude of mutant mice, the state of gene therapy for epilepsy, and an unseeing gene of the month.
In this episode
00:57 - Dr Julian Knight - Genes and immunity
Dr Julian Knight - Genes and immunity
with Dr Julian Knight, Wellcome Trust Centre for Human Genomics, Oxford
Kat - From the moment we're born, we're exposed to millions of bacteria and viruses in the world around us. Most of the time they don't make us ill, but if they do, most of us can fight off the infection. But for some people, it's a different story, and they can become desperately sick. I spoke to Dr Julian Knight from the Wellcome Trust Centre for Human Genomics at the University of Oxford, to find out how genetic studies are helping to reveal more about these differences.
Julian - A classical approach that people have used is to look at the very rare examples of patients who have a specific genetic difference that predisposes them to a particular disease. For example, with very young children, they may be born and have genetic differences that result in severe primary immune deficiencies where they're very vulnerable to infections. The other way of thinking about the genetic contribution to immune related diseases is with common diseases which might include infections or autoimmune diseases like rheumatoid arthritis.
We now have the genetic tools to actually try and look across the whole genome and look for what are often several different genes and genetic differences that may be contributing to the same disease. That's really been driven by new technologies that have become available to us within the field of human genetics such that, rather than focusing on a single gene, we can now look at many thousands of different genes and indeed, many millions of different genetic differences.
Kat - I guess, to draw an analogy it would be a bit like the more severe diseases, you look at a car and the wheel has fallen off and you can see that whereas - I guess what you're trying to do with the population saying, well, some people are more like a Ferrari, some people are more like a Fiat Panda. How do you even go about classifying all those huge number of differences?
Julian - An approach that's been successful over the last 4 or 5 years is to take a set of patients - so, you define a particular disease phenotype, so the condition with which the patients are manifesting, and then you define a set of individuals who are controlled, who don't have your disease, and you try and look for the genetic variants that are in one group, but not present in the other. By doing that, you can try and build up a map of where those genetic differences are lying and what particular genes they may or may not be influencing. Because it may be that the genetic difference is arising within the coding sequence of a gene and that would change the structure and the function of the protein that's encoded by the gene. And that might be something that you can very readily detect. Or it might be that it's a genetic difference lying outside of the gene within the regulatory sequence that controls how much protein we produce. Although those are more subtle, they're perhaps more common and very much more important in these complex disease traits than we'd initially anticipated.
Kat - So, what do we know so far about the kinds of genes that are involved in regulating our immune system and some of the differences between people and their immune systems?
Julian - We've got some evidence that goes back a long time in terms of genetics research - we're talking 40 or 50 years now - that we've understood that genes within a region on the short arm of chromosome 6 called the major histocompatibility complex. We know that genetic differences in that region are critical to encoding proteins that present foreign antigens to the immune system and that's critical in our response to infection.
Kat - So, this is like picking something up and going, "Look at this immune system. Recognise this and fight it."
Julian - Absolutely, and that takes quite complicated machinery to break up the protein loaded on to what we call a presenting molecule that can then be taken to the surface of the cell and other cells within the immune system can recognise that and initiate what we hope is an appropriate immune response to deal with that infection. In some instances, we think that that process goes wrong, and that can lead either to an excessive immune response which might actually be harmful to us. Or indeed, it might be that the body starts to fight itself and that can lead to autoimmune diseases. But now that we can look at the rest of the genome, we know that outside of the MHC, there were also important differences between people and patients in particular that contribute to developing the risk of disease
Kat - When we discover more about these genes, not only is this helping us to understand who's more vulnerable to certain diseases or not, would this explain why some people for example, seem to get flu all the time or some people seem to be very sickly and others are very robust?
Julian - Absolutely. So, over in the hospital, we see elderly patients who are very dependent and are coming perhaps from a nursing home with a severe infection. And despite the pessimism that they might be in some of those situations, these patients can be remarkable and they can survive. We know that people differ in how well they're able to respond to infection, how appropriately they can respond to infection, given that this is still a major killer in our society in terms of severe sepsis - the mortality rate on intensive care units is still of the order of 20%.
Understanding this very basic biology in terms of why some people are able to survive and overcome severe infection I think is potentially very important because it can lead us into potentially new therapeutic avenues, drug targets for example, or indeed, being able to better understand where and who we should be targeting therapy for. It may be that possessing particular genetic differences means you produce more or less of a particular protein, critical to an immune response. If we can understand that both at a population level and in the lab, then hopefully, we can go some way forward to understanding the genetic basis of these diseases.
Kat - The knowledge that you're gaining now about how people's immune systems respond in different ways to diseases and to things like sepsis, how close are we seeing this genetic knowledge actually coming into the clinic and benefiting patients?
Julian - Well, I think that we're perhaps still 2 or 3 years away from really understanding with confidence what these genetic contributors are to diseases such as severe sepsis. We have been able to identify in specific patients who have rare inherited defects or problems with their immune response particular genes and that can be very helpful in terms of making a diagnosis and guiding therapy. But it may be that if we can understand these genetic causes better and predispositions, then perhaps there are some patients who might benefit from therapies who we can target specifically to use such therapies. By only targeting those patients who are going to benefit, we can avoid the downside of these sort of therapies, which paradoxically increase your risk of infection in some cases.
So, I think that there is a hope that by using genetics, we can more rationally use existing drugs, we can identify targets, whereby we can develop new drugs or use existing drugs in new ways. We may be able to reduce what is really, still a devastating disease whereby, we're faced with high mortality rates despite best care in terms of the intensive care units and available antibiotics and so on. I think that's what motivates a lot of people who were interested in trying to use these new genetic approaches in a whole range of different diseases.
Kat - That was Dr Julian Knight from the Wellcome Trust Centre for Human Genomics.

08:56 - Many new mouse mutations
Many new mouse mutations
with Prof Karen Steel, Kings College London/Wellcome Trust Sanger Institute
Kat - Researchers at the Wellcome Trust Sanger Institute have published the first batch of results from an ambitious project aiming to systematically knock out, or delete, every single gene in mice, one at a time. Not only have the results provided a whole bunch of scientific surprises, but the scientists are creating an incredibly valuable resource for biomedical researchers around the world. Here's Professor Karen Steel, lead researcher for the project, to explain more.
Karen - The Sanger Institute several years ago, set up a project called 'The Mouse Genetics Project' and the aim of this was to develop a large panel of new mouse mutants, each of them with a single gene targeted or mutated. Then with these mutants, we put them through a screening process to pick out a wide variety of different signs of different diseases, and also, variations in phenotype, normal aspect of the function of the mouse.
Kat - What sort of diseases were you looking at?
Karen - The whole range, ranging from hearing and deafness which is my personal interest, to metabolism, to vision defects, to general developmental defects that could affect the development and morphology of any part of the body, and fertility and viability, so a very wide range of things.
Kat - What did you find when you looked at all these mice doing all these things?
Karen - Well, we've just published the first analysis of the first 250 mouse lines that we've generated. One of the things that we found from that was that some of those genes were known, well known, and other genes were completely unknown and nothing had been published about them previously. What we found was that we were just as likely to find some phenotypes and aspect of the function of the mouse in these new genes, these novel genes that hadn't been described properly before other than their sequence compared to the well-known genes.
So, what that tells us is there's a lot of genes out there in the genome that nobody is paying attention to or studying. Really, they are likely to be a very rich resource for finding associations between genes and between function and particularly, between genes and diseases.
Kat - So, we need to look outside the usual suspects and start looking at all these other genes as well.
Karen - Absolutely, yes. Of course, some of the genes that we studied were ordinary and there have been papers published about them. But by doing systematic screen, putting every mouse line through the same battery of tests, we found new aspects of their phenotype that hadn't been published before, that haven't been found before. So, it gives them that fuller picture of what is likely to be going wrong as a result of a mutation in specific single genes.
Kat - So, you've looked at 250 different mouse mutations so far. How many more have you got to go and how long is that going to take?
Karen - Well, there are 20,000 genes approximately in the mouse genome.
Kat - A long time!
Karen - So, it's going to take a while. At the Sanger Institute, we've generated over 900 mouse lines so far, so we're getting along quite well. It's still a long way to go before we get all of the genes. As we generate those mouse mutants, and breed enough mice to put through the screening, then the data from those new lines are going onto the website.
One of the really key aspects of this resource is that it's a public resource. So, anybody can go and look on the website find all of the data about the phenotypes, the characteristics of these different mouse mutants. Scientists and clinicians can also get access to the mouse models themselves so that they can get that mouse into their own laboratory and do their own specialised tests because they're the experts in their own disease areas. And so, they're likely to be able to do a much more detailed analysis than we can do as part of a screen.
Kat - That was Professor Karen Steel - now at King's College London but previously at the Wellcome Trust Sanger Institute where this research was done.
13:03 - Tracing lung cancer
Tracing lung cancer
with Prof Charlie Swanton, Cancer Research UK London Research Institute
Kat - Moving from mice to humans, this month the charity Cancer Research UK announced their own ambitious project, sequencing the genes in tumour samples from more than 800 patients with lung cancer and tracking how they evolve over time in response to treatment. It's hoped that the study, called TRACERx, will revolutionise scientists' understanding of the genetic faults that underpin lung cancer, and reveal more about tumour evolution and so-called "tumour heterogeneity" - the fact that cancer cells within a single patient can have different genetic makeups. Here's the project's leader, Professor Charlie Swanton, talking to Cancer Research UK's Greg Jones.
Charlie - So, we're going to be looking at how lung cancers, principally non-small cell lung cancers change over time and how their spatial and temporal variation gives us some insights into potential new therapeutic strategies for patients. So, what we've realised from our work over the last year or two is that tumours are not just single entities but they're composed of multiple different subclones that maybe intermingled or spatially separated. What we're also realising that tumours are evolving over time. So, there's a spatial and temporal aspect of tumour biology that many of the sequencing approaches we've been taking so far have largely not taken into account. Principally because of the cost involved and also, I think the awareness now that tumours are markedly more heterogeneous than perhaps we had imagined initially.
So, the idea with TRACERx, which stands for Tracking Cancer Evolution Through Therapy, is that we ask patients with primary non-small cell lung cancer to consent this study which would enable us to acquire any tumour material that is surplus to pathology requirements, following surgery - so, these are patients with primary operable lung cancer - and then we will subject each tumour to multiple sequencing approaches to identify what are the shared mutations in every region and what are the diverse heterogeneous mutations in every region.
If the patient is unfortunate enough to suffer recurrence of disease or metastatic disease, we'll ask if the patient would kindly consent to a further biopsy so that we can then compare the biopsy sites to of metastatic disease. So, the original primary to ask the principal question, how has the disease changed over time, to better understand the biology of metastatic disease, to better understand resistance to therapy, and ultimately, to come up with better clinical approaches to treat this disease to stop this from happening.
Greg - Can you explain why this is going to be such an enormous undertaking in terms of the amount of data you're going to generate?
Charlie - So, we're sequencing tumours from 850 patients, but we're not sequencing just one tumour. We're sequencing up to 6, 7, or even 8 coding exomes from each tumour during the disease course. Now, a coding exome has about 50 million base pairs and we're sequencing at a depth that is relatively unprecedented in these studies - at 500 so-called X coverage. And so, if you take that into account, along with the number of tumours we're sequencing in a number of regions we're sequencing within each tumour and comparing primary to metastatic states, we're talking a need or requirement of probably 6 petabytes worth of data storage. That's the equivalent of probably sequencing 42,500 whole genomes at one X coverage in order to really get to grips with the diversity within a single tumour.
Kat - That was Professor Charlie Swanton from the Cancer Research UK London Institute.
16:24 - Is fatness in the genes?
Is fatness in the genes?
Scientists at Boston Children's Hospital have discovered that mice carrying a faulty version of a gene called Mrap2 gain weight, even while eating the same amount as their genetically normal counterparts. The gene is involved in a signalling pathway in the brain that increases energy burning while decreasing appetite.
Writing in the journal Science, the scientists suggest that mice with a faulty version of Mrap2 seemed to be hanging on to fat rather than burning it. Interestingly, the animals gained even more weight on a high fat diet, compared to regular mice. Faults in the human version of the gene have also been found in severely obese people, although they're only found only in less than 1 per cent of the obese population, suggesting that MRAP2 could play an important role in weight control and energy balance.

17:09 - Genes and cholera
Genes and cholera
US researchers studying the genetic makeup of hundreds of people in Bangladesh have tracked down key regions of the genome that could be responsible for making some people more susceptible to infection with the bacteria that cause cholera, publishing their results in the journal Science Translational Medicine. The genetic regions harbour genes involved in the immune system and regulation of fluid loss, among others.
The next step is to pin down the exact genes responsible for the differences in susceptibility and find out what they're doing. Cholera is still widespread in many parts of the world, and can kill within hours. The scientists hope their discovery will pave the way for better vaccines and treatments, to help cut the death toll of cholera outbreaks in future.
17:50 - Chemicals reprogramme cells
Chemicals reprogramme cells
Chinese scientists have made an important breakthrough in stem cell technology, showing that they can convert adult cells into stem cells, with the capacity to become any type of tissue, using just chemicals rather than genetic engineering. Back in 2006, Nobel prize-winner Shinya Yamanaka showed that adding a cocktail of genes to adult cells could take them back to a stem cell state, known as inducible pluripotent stem cells. Now Hongkui Deng and his team have managed to get the same effect in mouse cells using a handful of chemicals.
The researchers, who published their findings in the journal Science this month, are now trying to get the technique to work in human cells, although it's proving tricky. But if they can make it work, it could be extremely useful, as four of the compounds are already in clinical use. But for now, we'll have to wait and see.

18:54 - Prof Mike Levin - Childhood infections
Prof Mike Levin - Childhood infections
with Prof Mike Levin, Imperial College London
Kat - But now it's time to find out a bit more about genes and the immune system. Perhaps one of the best places to see the role of genetics in shaping the immune response is in childhood. All kids are exposed to a huge range of bugs - but only a very small proportion will get life-threatening infections, when most others will be unaffected. Even in developed countries like the UK, childhood infections are a big problem, and hundreds of children die from bacterial and viral infections every year.
Professor Mike Levin is Professor of Paediatrics and International Child Health and Director of the Wellcome Centre for Clinical Tropical Medicine at Imperial College in London. I caught up with him to find out more about the genetics of childhood infections.
Mike - So, as children are born and grow through childhood, they're exposed to an absolute zoo of different bacteria. A baby is born from a sterile womb and in minutes of coming out into the world, they are colonised by all sorts of bacteria that are in the environment. Now mostly, as the child develops, they come to no harm. Their immune system keeps the bacteria where they should be - that's on the skin, on the mucosal surfaces, inside your gut - but 1 in 20,000 or 1 in 50,000 children, the bacteria invade from the mucosal surfaces - the nose, the throat, the bowel - get into the bloodstream and cause life-threatening infection.
That process, so why does one child come down with the infection and not others is partly controlled by the virulence of the bacteria. So, there seem to be some bacteria that are worse than others. So, there are certain bacteria which are more likely to invade if they meet a young child, but there are other bacteria that are completely harmless to 99.9 per cent of the population. 1 per cent will come down and be critically ill.
Kat - Can you tell me a bit more about the particular study that you're involved in looking at how genetics influence childhood disease?
Mike - We were very fortunate to be awarded a major grant which is funding a study EUCLIDS which stands for the European Union Childhood Life-threatening Infectious Diseases Study. Essentially what it is, is it's recruiting children who were admitted with infection from multiple hospitals in the UK as well as from multiple hospitals in Holland, Spain, Austria, Germany and probably soon, Switzerland, as well as the Gambia in West Africa.
This is enabling us to establish very large numbers of patients who are very carefully studied and the grant also provides a funding to apply very sophisticated genetics including sequencing and genome-wide association methods to this very large cohort. We hope that the study will provide very good information on the genetic basis of a range of different childhood infections including Staphylococcus infection, Streptococcus infection, Pneumococcus, Meningococcus, and Salmonella which are very important childhood pathogens.
Kat - Where are we heading in terms of turning this into benefits for children and actually improving child health?
Mike - I think there are a number of reasons why unravelling the genetic basis of how the immune system responds to bacteria is going to help in the future. The first is that obviously, for very severe defects, if you know what the gene is, you can test for it and you can offer counselling and treatment for those that are affected. For example, there are a set of gene defects in a gene and a protein in the blood called complement which you need to kill the Meningococcus bacteria. If you found to lack this protein complement, then you're going to be at lifelong risk of coming down with meningococcal disease. We can treat those patients by giving them preventative antibiotics. So, that's the simplest example if we know what the gene is and we can offer prevention.
The more sort of subtle example is that if we know how the immune system works, to stop getting an infection - which the gene defects are often a clue to - then we know what sort of immune effect we need to stimulate with a vaccine. So, understanding how the immune system works through the genetics can improve understanding of how to develop vaccines. I suppose the third way is that it looks like, not only do genes control who gets the infection, they also control how bad it is and what the outcome is.
So, there are some people who will get bacteria in the bloodstream and they'll come in to a hospital and receive a course of antibiotics, and go home and are cured completely. There are other children that come in become devastatingly ill, and may die or lose limbs in a matter of hours. It looks as if those different responses are also genetically controlled. Again, if we understand what is going to make one child become catastrophically ill, another one become milder, then one can develop strategies to improve the outcome. I think again, the genetics is a powerful clue to why some people do badly and others do better.
Kat - It seems only in a matter of maybe 100 years, we've gone from many, many children dying in childhood from simple infectious disease to now a point where, certainly in developed countries, many, many children are saved. Where do you think we're going to be heading in the next 10 years? Where would you like to be?
Mike - I think, hopefully, the reduction in childhood mortality from infection that has been seen in the developed world will be extended to the less resource-rich areas of the world. So, I think we would hope that the advances in prevention of infection through vaccination hygiene and treatment which has meant infection causes fewer deaths in Europe, the United States, will start happening in Africa and Asia. So, I think that's a big goal. Although infection has declined as an important since the earlier part of the last century, it's still there.
So, those of us working in paediatrics services continually see children admitted critically ill with life-threatening infections like meningitis, septicaemia, pneumonia, osteomyelitis. Trying to work out how to improve the outcome for those that are infected, trying to work out how better to prevent infection is going to remain a research challenge. So, all the research is still needed because many of the infections are still with us. There's still a major problem - although things are very much better than they were 100 years ago - there's certainly no room for complacency. A large part of the workload of paediatric services, paediatric intensive care, is still infectious diseases.
Kat - That was Professor Mike Levin from Imperial College.
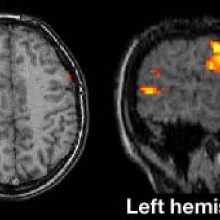
Is gene therapy for epilepsy in the clinic yet?
Kat - Now it's time to look at your burning genetics questions, with the help of Naked Scientist Martha Henriques.
Martha - Listener Richard Salmon wanted to know whether the experimental epilepsy group at UCL, working on a gene cure for seizure disorders have progressed to human trials. Their study, published last November in Science Translational Medicine, showed that introducing a genetically modified virus to a brain region which generates the epileptic seizures suppressed excitability of that region, significantly reducing the effect of seizures. Stephanie Schorge, an author of that study at UCL told me that the group's application for human trials has been submitted. She explained that the first human trials for this viral treatment will be in patients who could have the current standard treatment for this type of epilepsy in which the brain region generating the seizures is surgically removed. Stephanie Schorge...
Stephanie - We give maybe five patients our treatment and then if the seizures get better, great! We're happy. If the seizures don't get better, we could have that secondary treatment of having the tissue that was injected with the viruses causing seizures and having that cut out. So in a way that's a safety thing in humans that we're injecting tissue that we know otherwise would have been removed anyway. And then if it proves safe and effective in these five patients, we go to our next population which is our ultimate population for patients who have seizures coming from parts of the brain which we know cannot be cut out. And these patients who have seizures rising say, from the motor cortex which controls an arm, if we can show that in animals injecting this virus has no effect on movement, then we think that if we inject the same virus into the human cortex it's also unlikely to affect movement.
Martha - Thanks to Stephanie Schorge for that answer and to Richard Salmon for his question.
Kat - If you've got any questions about genes, DNA and genetics you'd like us to answer, just email them to genetics@thenakedscientists.com, tweet us @nakedgenetics or post on our Facebook page.
28:33 - Gene of the month - Eyeless
Gene of the month - Eyeless
with Kat Arney
And finally, our gene of the month is Eyeless - a fruit fly gene that has close relatives across the whole animal kingdom. Flies with a faulty version of Eyeless, as you might have guessed, fail to develop eyes. This is because the gene is a kind of master switch that sets up the whole eye-forming process in the developing embryo. The human version of Eyeless is called Pax6, and children who inherit a faulty version are born blind. There are also version of Eyeless in fish, highlighting its importance throughout evolution. By studying Pax6 and Eyeless, researchers are starting to understand how the instructions to make eyes are laid down, as well as gaining important insights into hereditary blindness.
- Previous Chitin: Chemistry in its element
- Next Shooting stars










Comments
Add a comment