In this issue of the eLife podcast we discuss how flatworms can grow new heads and tails, how photosynthesis has evolved over time, social interactions between mice, the properties of "undead" cells, and interactions between steroids and genes.
In this episode
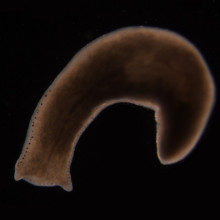
00:49 - How flatworms regenerate tissue
How flatworms regenerate tissue
with Peter Reddien, MIT
When a part of the body of some organisms is lost, they can grow a replacement, but what controls this and how does it differ from when individual cells wear out in a tissue and needs replacing? Worms - it turns out - might be able to help us. Peter Reddien spoke to Chris Smith... 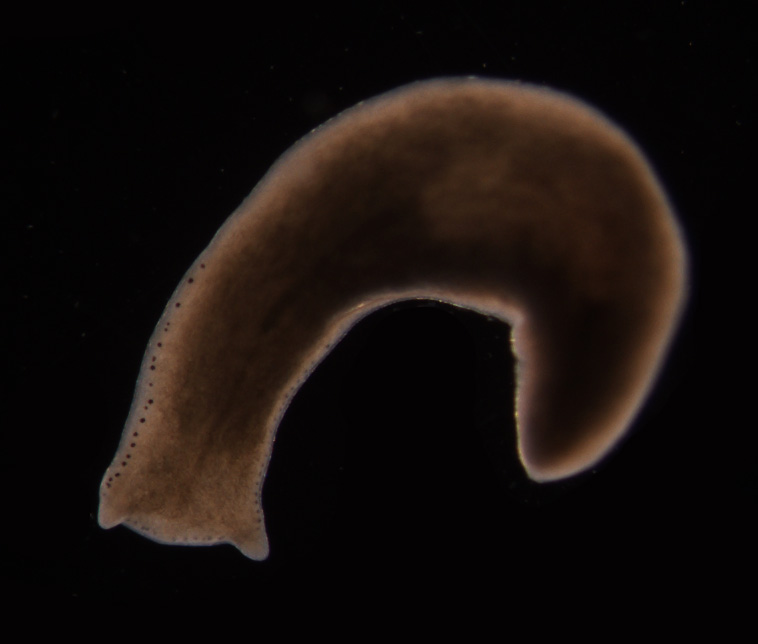
Peter - My name is Peter Reddien. I'm an Associate Professor of Biology at MIT and a member of the Whitehead Institute. The problem was how regeneration initiate it. In particular, we were looking at this process in an organism called planarians. Planarians are flatworms. These are animals that are capable of regenerating anything. They can regrow new heads, new tails or entire organisms from tiny fragments of the body. One of the questions that's central to understanding regeneration in any organism is, how does it start? You wound an animal and then what happens to bring about all of the events of regeneration?
Chris - So, how did you attack that problem?
Peter - We attacked this problem by investigation of genes that are activated at wounds. We had found dozens of genes that are turned on by injury. Through investigation of a set of these genes, we found one required for regeneration. So, we did this with wound induced genes, inhibiting them one by one using RNA interference and found a particular gene called follistatin that when inhibited, cause a block in regeneration.
Chris - What was the nature of the injury that you were testing out on these worms?
Peter - Well, the nature of the injury initially was complete head removal and tail removal. This leaves a middle piece that's capable of regenerating a head on one end and a tail on the other. What happened was quite remarkably, these animals completely failed to regrow new tissues. What made this particular defect catch our eye was the fact that despite failing to regenerate, these animals in which we had inhibited the gene follistatin were capable of living and capable of replacing aged cells during natural tissue turnover. What I mean by that is that there's a population of cells that are involved in normal adult life in planarians and involved in regeneration. This population is called the neoblast population of cells. We know from prior work that within the neoblast are pluripotent adult stem cells. So, these pluripotent stem cells are dividing and producing new specialised or differentiated cells. For example, let's say an epidermal skin cell gets old then a neoblast will replace it with a new young one. A gut cell gets old, neoblast will replace it, and so on. That process of tissue turnover is happening constitutively and is essential for life. This process happens normally in the animals lacking normal follistatin function.
Chris - So, the fact that you could interrupt this follistatin signalling and stop the regeneration of the gross part of the body, indicates that that sort of regeneration is a different process from the tissue maintenance regeneration that's just repairing and replacing cut out cells at a tissue level.
Peter - That's exactly right. The difference in the defect between tissue turnover where everything was fine and regeneration really caught our eye, suggested that this gene may have a really unique and permanent role in regeneration specifically.
Chris - How does a tissue discriminate between the removal of a big chunk of the body, like the head, and the loss of a few cells just due to oxidative damage, you wet something that you didn't particularly agree with?
Peter - How they distinguish between the absence of a cell that's gotten old and the absence of a set of cells from injury is still a mystery that is unsolved. But what we know is that when there is an injury that has removed a substantial amount of tissues, injuries like that trigger a sequence of events that are specific to regeneration. There's particular events with the regenerative cells called neoblast, such as the migration of these cells to the wounds or do they continue division of these cells at wound sites that only happens when you actually remove something. We call the sequence of events that happen only in injuries where you've removed substantial tissue, the missing tissue response. And it is these events that follistatin is required for.
Chris - Looking downstream of follistatin, what does it actually appear to be doing to the target tissue once its produced under these traumatic injury situations?
Peter - So, follistatin is induced and what is required for are a series of events that are specific to regeneration. So, it's required for neoblasts - the regenerative cells - to specialise into replacement cells. We looked at this using eye regeneration and neoblast normally specialise into eye progenitors following head removal. This did not happen following inhibition of follistatin and amputation. In addition, neoblasts display sustained proliferation at wound sites and cases in which they need to regenerate substantial tissue. This was also defective in follistatin RNAi animals. Finally, there's increase in death of cells throughout the body that occurs when substantial tissue has been removed by injury. This process of death or apoptosis can be readily visualised with molecular assays and it failed to happen in the absence of follistatin.

06:54 - The evolution of photosynthesis
The evolution of photosynthesis
with Julian Hibberd, Cambridge University
The process of photosynthesis goes back over 3 billion years. But in just the last 30 million years or so, a new and dramatically better form of photosynthesis has appeared. But it requires a considerable number of changes within plant structures to sustain it. Speaking with Chris Smith, what Cambridge plant scientist Julian Hibberd wanted to know was, in what order (if any) these changes needed to have occurred to make this so-called "C4 photosynthesis" feasible.
Julian - Photosynthesis is the synthesises the mechanism by which almost all life on this planet is maintained because it's the system by which CO2 is converted into sugars and carbohydrates. It provides our basic food stuff. Historically, it provides our fuel because a lot of fossil fuels were laid down by ancient plants. And so, we're interested in how a particular flavour of photosynthesis evolved about 30 million years ago because it's actually more efficient than the ancestral version of photosynthesis.
Chris - So, tell us about the two types of photosynthesis. What are they and how do the differ?
Julian - The ancestral version of photosynthesis, we think, evolved in bacteria about 3.6 billion years ago and that uses an enzyme which fixes the CO2 into sugars. That enzyme never had to distinguish between the CO2 molecule that it now uses, and oxygen, which is now present in the atmosphere. The reason for that was, those ancient bacteria were in an atmosphere without oxygen; but, later on, some more complex bacteria evolved which started to pump oxygen out into the atmosphere. As a consequence, the photosynthetic organisms today have to distinguish between carbon dioxide - CO2 - and oxygen. At the molecular level, they are actually quite similar. And so, this poor protein, which is meant to fix CO2, some other time makes mistakes and uses oxygen instead.
Chris - That presumably reduces the efficiency of the process?
Julian - Yeah and the efficiency is reduced the most in the tropics and subtropics, because there's a correlation between the specificity of the protein and its ability to fix CO2. As you get warmer, it fixes CO2 less efficiently.
Chris - So, how did evolution tackle that?
Julian - So, evolution has tackled it actually on many occasions. In the land plants, we think at least 60 different lineages of land plants evolved the derived version of photosynthesis that I work on and there are other lineages which have evolved other mechanisms to cut to concentrate carbon dioxide around this photosynthetic protein. We've become interested in how - what is actually a really complicated trait kind of evolved so many times and the evidence at the moment is it's over 60.
Chris - When this trait evolved, what sorts of things have had to change?
Julian - In a C4 leaf, there are changes to the structure of the leaf itself, to the leaf anatomy. There are changes to structures within the cell. So, cell biology changes and there are also really distinct changes to gene
expression and the spatial arrangement to photosynthesis within those cells in the leaf. So, it's really quite a complicated suite of alternations which have to occur.
Chris - Your question is, in what order did those things crop up? Excuse the plant pun.
Julian - Exactly. So, we're interested in whether specific traits occur early along that evolutionary path to see for photosynthesis or late or actually, whether they can be flexible, whether they can occur in some lineages very early on and some lineages much later on. But you still get to the final phenotype at the end.
Chris - So, how did you do this?
Julian - What we did was we took about 73 different plant species. Some of which used the ancestral C3 version of photosynthesis. Some used this derived C4 version of photosynthesis. Some remarkably which have characteristics of both of those types of photosynthesis but without the full syndrome. We mapped onto those species which trait were visible in each. And so, we characterise them as either having a set of 16 C3 traits. So, that species was completely C3, completely ancestral in terms of photosynthesis or another species might have 16 C4 traits. So, it will be completely derived. In the intermediate species, we'd have a mixture of some C3 and some C4 traits.
Chris - I suppose if you look at the evolutionary timeline separating the ones which are fully C4 and those that are fully C3, you can ask, where along that timeline some of these traits must've occurred.
Julian - Exactly. So, we've estimated that some traitsare very likely to occur early on in the transition from C3 to C4 photosynthesis and conversely, some always come in at the end. But there are a few which seem to be bimodal. So, they have different distributions. Sometimes they come in late and sometimes they come in early which implies that there are many flexible routes for a plant - a photosynthetic organism - to move from this ancestral version of photosynthesis to this more efficient derived version of photosynthesis.
Chris - Putting all of your findings together, what then is the bottom line?
Julian - So, what we discovered was that this complex system which has evolved multiple times is likely to have evolved multiple times because there are many ways of getting to the same end point. So, 'many roads lead to Rome' I think is a phrase which could be used to describe our system. This very complicated version of photosynthesis seems to have got there on multiple occasions because there are many ways to reach that final goal. The other thing that we infer from our results is that the initial environmental drivers for evolution of C4 photosynthesis is unlikely to have actually been improvements in photosynthesis itself, but more likely to have been changes in the ability of the plants to use relatively low water supplies or something like that.
13:34 - Social interactions in mice
Social interactions in mice
with Alon Chen, Weizmann Institute, Israel
Chris Smith spoke to Alon Chen from Israel's Weizmann Institute has developed a computer system to log automatically how the animals interact with each other. Commenting on the work, Cambridge neuroscientist, Hannah Critchlow...
Hannah - This new system has taken 5 years to develop and may save hours of scientists time on consuming data analysis. Inspired by disco lights, it works by painting the fur of individual mice with different coloured UV paints, illuminating them with the UV lamp and then recording the movements of the nocturnal mice by night. A computer system then takes the video to movements and generates tens of thousands of rows of data per hour of recorded footage and translates these into predictive behaviours such as grooming, sniffing, feeding together, exploring or simply avoiding each other . By providing it a system for analysing complex social interactions between large groups of mice, this could pave the way to a greater understanding of disorders associated with altered social interaction, such as autism. Researchers can tweak the genes or environment of mice and then relatively quickly see how it affects their social behaviour. The scientists have firstly applied this technique to find out how enriched environment affects how mice interact with each other. They found that adolescent mice that have grown up in a stimulant rich environment full of games and toys were much more territorial. So, they didn't share their food as much. When they did interact, they worked in pairs rather than as a larger group. Mice growing up in more Spartan conditions were much more community-minded. So, the scientists suggests that this finding in mice ties in with the common belief for humans that our modern stimulation filled environment encourages individualistic behaviour whilst simplest surroundings give rise to a more developed community life. I spoke to one of the authors of the study about the implications of this result, Professor Alon Chen...
Alon - So, we have in our case, a very specific study in which we just provided the mice during development, during - in a way - adolescence, more stimuli. In this case, it became in a way less social because each one have much more resources in a way that develop its own territory. And therefore, it probably need less the surrounding compared to a basic group. Again, you can think about human societies and which have much more reach type of resources versus the other one whether they are less or more individuals. So, it's easy to see equivalent system in humans.
Chris - Alon Chen and before him, Hannah Critchlow. Alon told Hannah that since publishing this work, he's received requests to share the system from researchers all over the world. Under certain circumstances including during development, large numbers cells kill themselves and it's clear that when one cell commits suicide, it can persuade other cells to follow suit, but how?
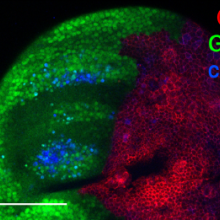
16:43 - Cell death at a distance
Cell death at a distance
with Hermann Steller, Rockefeller University
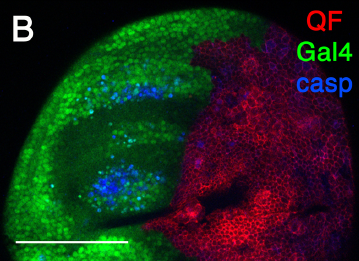 Under certain circumstances including during development, large numbers cells kill themselves and it's clear that when one cell commits suicide, it can persuade other cells to follow suit, but how? Hermann Steller spoke to Chris Smith...
Under certain circumstances including during development, large numbers cells kill themselves and it's clear that when one cell commits suicide, it can persuade other cells to follow suit, but how? Hermann Steller spoke to Chris Smith...
Hermann - My name is Hermann Steller. I'm a Professor at the Rockefeller University in New York and also, an investigator of the Howard Hughes Medical Institute. It's been typically considered that cells undergo apoptosis by themselves that they make the decision between life and death and then carry out this programme - cell death mechanism - to self-destruct. And the current study actually shows that these cells are very active in influencing each other in their life and death decision. Cells that die can still lead other cells to die as well. This is significant because both in normal developments and in certain disease situations, large groups of cells die in a somewhat coordinated behaviour and the signal to die can be heard amongst the population and coordinated suicidal behaviours achieved, a little bit like lemmings, all jumping over the cliff.
Chris - So, some kind of signal originates from a cell or group of cells that have become destined to die and other cells, what in the near vicinity die, or does this signal potentially work over longer distances?
Hermann - Well, the surprising thing in the study was that the signal can work over considerable distances. A group of cells in one place can signal many, many cell diameters away to another group of cells that it's time to die. Now, I should say that initially, apoptosis has been seen as something where the only thing that the dying cell signals to environment as an 'eat me' signal so that the corpse is removed. Subsequently, a number of labs have shown that cells, it can actually release signals to stimulate cell division. That seems to make sense in a situation when a cell is lost that you want to replace. So it's, "I'm dying. Please replace me" signal. So, in this context, it was very surprising that when there's a strong stress on a tissue that the dying cells can release signals that travel over considerable distances to stimulate more cell death.
Chris - How did you actually discover this?
Hermann - Well the post doc in the lab studied the signalling of dying cells in the developing drosophila wing. He generated a strong cell death situation in one piece of the wing and then she saw to her surprise that in a region quite far remote that there was cell death at the distance.
Chris - How did you find out that it wasn't just the fact that lots of cells were dying and there was some kind of necrotic effect if you've got lots of dead tissue, this might be inducing the death? How did you prove there had to be some signal made actively and sent from cells that are destined to undergo apoptosis which was triggering other cells remote to die?
Hermann - So, we use a system where we, on the one hand, induce cell death with key signalling molecules to activate cell death programme. But then we can arrest the execution of cell death with an inhibitor of the proteases that are responsible to carry out cell death. So, in this system, the cell basically thinks it's going to die, but it's prevented from carrying out the real destruction. So, these are what we call 'undead cells'. They are really cells that think in a way these cells are dying, but they stay alive. As a result of that, these cells also signals for much longer, virtually indefinitely and allow us to look at the signalling context. So, there's really no cell death and no tissue loss occurring but still, the phenomenon of inducing death at a distance is observed.
Chris - So, you programme these cells to think they're dead, but they remain viable and pump out whatever the signal is. You see corresponding death in other cells in other bits of the embryo. So, how did you then track down what the chemical was that was doing that signalling?
Hermann - We had reasons to suspect that two candidates signals. One is the Jun Kinase pathway, the JNK, which has been implicated in signalling decisions in cell death and ligands with this pathway called eiger which is related to tumour necrosis factor conserved rodent skin, in both insects and in mammalian systems.
Chris - How did you then explore what those were doing?
Hermann - Well, we again could take advantage of the powerful genetic methodologies in drosophila to inactivate these pathways. So very strictly, we just take away the gene that encodes eiger and then ask if we would stop this apoptosis, induced apoptosis. Specifically, we were able, not just to remove eiger throughout the entire tissue, but we could specifically eliminate eiger function in the cells that would undergo apoptosis and that would completely abrogate the induction of apoptosis at the distance, even though these cells at a distance still have normal eiger function. This really shows that it is eiger required in the dying cells as a signal to travel and induce apoptosis at a distance.
Chris - But all around my body every day, there are billions of cells committing suicide in this way, so why don't I just unleash this dramatic domino effect of apoptosis and all my tissues self-destruct? Something must be opposing the apoptosis induced apoptosis phenomenon.
Hermann - Yeah, that's an excellent question and something where we really don't have very complete answers. it's through that small amount of apoptosis will actually lead to a compensatory response where the dying cell stimulates growth and regeneration. Only when there's sort of a large scale of death is apparently to signal strong enough to kill. But there must be protective mechanisms because even in the systems we've studied, we've seen cells in between of being affected. So, there's a different sensitivity that cells have and there must be a complex regulation to protect against the unwanted apoptosis induced apoptosis.
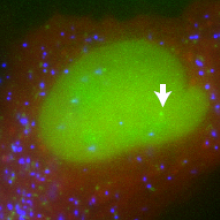
23:26 - Studying transcription in single cells
Studying transcription in single cells
with Dan Larson, National Cancer Institute
Looking at large populations of cultured cells, it appears that when a stimulus is applied, genes are turned on and gene products are made continuously and in proportion to the intensity of the stimulus. But if you look at individual cells, a very different pattern emerges. National Cancer Institute researcher, Dan Larson spoke to Chris Smith...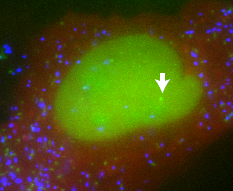
Dan - It turns out, and this is one of the punch lines of the paper, it seems like there are lots of ways for the nucleus to turn genes off. To turn the gene on requires lots of things to happen correctly. You need to recruit chromatin remodelers, you need to recruit what's called the basal transcription machinery. And we want to know how much of this one particular element was required for this gene to turn on. And the element that we looked at in this case was a steroid. We can quantitatively change the level of this steroid and we can look at the level of single RNA being transcribed to see precisely when transcription starts and how many RNAs are made.
Chris - So, the sort of premise here is that we know that when cells receive some kind of signal or they respond to some stimulus, the gene activity can be changed. Genes can be turned on.
Dan - Correct.
Chris - But what we're not so clear on is how the cells respond in terms of how much RNA, how much gene transcription goes on, in response to any given stimulus.
Dan - Yes. One of the ideas to emerge in recent years is that most RNA is not made in continuous levels. When you look in a population, you see this nice smooth increase in the amount of RNA may have the function of the amount of the upstream signal present. But when you look at a single cell level, RNA is transcribed in these short bursts of transcription. Punctuated by very long pauses where no RNA is made.
Chris - If we were to zoom in on the genome at the time that's happening, does that reflect assembly of transcription machinery or something which is why everything is there, all the ducks are in a row, it gets transcribed that then things stop while it resets itself or is there some other fundamental reason why you should get that very staccato, lots, nothing for a while, lots sort of phenomenon?
Dan - That's a good question. I would say in many cases, we don't know what's happening at the molecular level. What we know is that for example for this genome we studied in this paper, it transcribes for about 30 minutes and then it goes off for about 30 minutes even though the upstream signal isn't changing at all. Now, the timescales of this are suggestive possibly of chromatin remodelling events which may control some of the timing of this transcriptionional bursting or maybe due to the stability as you say of some of these factors at a promoter.
Chris - So, how did you try to get forward on this and get a handle on what was going on?
Dan - So, what we did is, we used a system that allowed us to visualise RNA in living cells. We make use of a system that mother nature has already used - the packaging of RNA into a bacteriophage. There's a bacteriophage capsid protein which binds a stem-loop and then that RNA containing a stem-loop is packaged into a particle. We have adapted that system and we use the same stem-loops and we encode them into our gene of interest. At the same time, we have capsid protein that's expressed in the cell and it's labelled a fluorescent protein. So once these stem-loops get made, the capsid protein binds and now, we can see this RNA in the microscope.
Chris - Wow! So, you can just count the particles that get made and that tells you how many RNA transcripts were coming off of that gene and see one time. What a phenomenal way of doing it. So, what have you learned?
Dan - So, what we've learned from that is that even when this gene is transcribing at saturation, there are long periods where no RNA has been made, again, due to this bursting phenomenon. When we increase the level of the steroid, the nuclear is set to respond to that steroid and a gene which responds to that steroid changes the frequency of this bursting but doesn't change the length of the burst and doesn't change the amplitude. So, what you can think of this is a toggle switch that's going on and off. Steroids control the frequency of this toggle switch.
Chris - So, what do you think are the implications of this? How does this inform or change what we thought before? Why is this important - the whole phenomenon?
Dan - I think one of the things that really comes out of this study is that we took a very well-understood physiological response. The response of a steroid responder gene to steroids. Biochemistry of these systems has been studied for 60 years and the dose response is one of the most well-characterised dose responses in pharmacology. It's a billion dollar pharmaceutical industry, understanding how steroids affect gene regulation. All of these studies are predicated on this idea that cells respond uniformly. That these dose response is due to the affinity of the receptor and the ligand. What we have shown is that when you look at a single gene level, you see something completely different. So, what we see is this complex frequency modulation which would never have been guessed based on ensemble studies and which suggests that this basic dose response mechanism may be way more complicated than we originally thought. It also raises the possibility that changing the dynamics of a ligand - in this case, the steroid - could lead to newer improved ways to use steroids as therapeutics.
- Previous Science Centre Showoff
- Next What is the Higgs boson?










Comments
Add a comment