Deciphering the Second Genetic Code
Interview with
Chris - Also in the new this week, researchers in Toronto and in Cambridge have made a major breakthrough in understanding how DNA works. More specifically, how the same gene can produce different gene products in different types of cells.
To tell us more is Yoseph Barash from the University of Toronto. Tell us first, if you could Yoseph, what's the problem you've actually been grappling with - what are you trying to solve here?
Joseph - Basically the problem that we were handling if I put it in one sentence would be to figure out how alternative splicing works. Of course, that wouldn't mean much if I don't explain what alternative splicing is and why it is important. So, I'll start off by starting with what people do know and people do know usually about the genes and that they're coded in DNA molecules, and many people know that scientists just about a decade ago mapped the human genome and they found about 20,000 genes altogether in the human genome. What people usually don't know is that the same gene can actually code for different genetic messages in the form of messenger RNA molecules. And these different messages can operate quite differently in the cell.
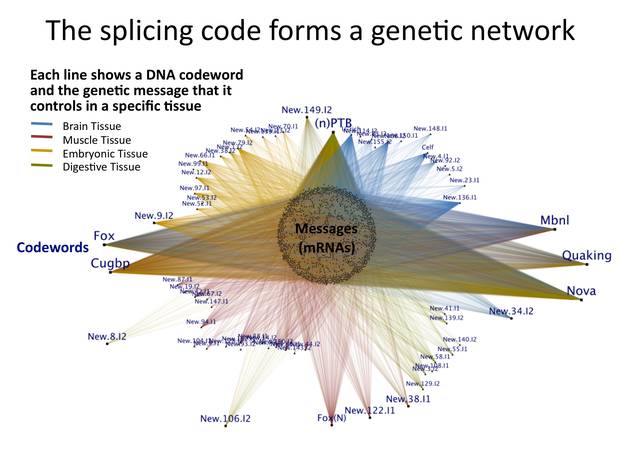 Chris - So in other words, in different tissues, genes which have the same genetic code can have a different effect by effectively chopping the gene up in a slightly different way, so it turns into a different recipe.
Chris - So in other words, in different tissues, genes which have the same genetic code can have a different effect by effectively chopping the gene up in a slightly different way, so it turns into a different recipe.
Joseph - Exactly. So instead of a one-gene one-product kind of model, we have a one-gene many products model and what we're trying to figure out is how this works. So, what is the code within the genetic code that tells the cell how, when, under what conditions, et cetera to perform these splicing variants.
Chris - So in a nerve cell, the same gene may do something completely different to a liver cell, but the big question is, how does it know it's a nerve cell or a liver cell and therefore to behave different?
Joseph - Exactly.
Chris - And how did you approach that?
Joseph - Basically what we did is interdisciplinary research that we started off by doing experiments and that was done at the Blencowe lab and we measured around 4,000 pieces, called axons, of genetic messages across 27 different mouse tissues. Then we analysed the data to figure out how these changes occur. So, the different inclusion or exclusion of these bits and pieces of the messages in the different tissues, how does it change? And then we went to the genome to figure out what is this code, what are the different components that determine these changes, so we can actually look at the genetic code and figure out if we just look at it, what would be the changes in say, brain versus liver as you suggested.
Chris - So in other words, by looking at many thousands of genetic sequences and doing this lots and lots of times in lots of different tissues, you can begin to tie together how a different gene gets cut up in a different way in a specific tissue, and then you can begin to work out what sequences are hidden in the genetic material that's making that happen.
Joseph - Exactly and that's the computer science part of the research - the machine learning part of the research.
Chris - So presumably, this is really important because what this will enable us to now do is when we want to do gene therapy on something, up until now, we've taken a very simple approach and said, "This gene turns into this product in a cell regardless of what cell type it is." So we just put the gene in and we'll get the product out. It hasn't always been as successful as we would've liked. Now, we're in a position to apply the discovery you've made which means that we can begin to ask, "Well, will this gene behave the way we think it will?" So presumably, your model will enable us to make predictions so that we can work out how genes will behave in different tissues.
Joseph - Exactly. So once you have that program, that model, then you can look at areas that you've never seen before, you've never measured in the original experiments, and use the program to tell you what's going to happen. You can also relate certain mutations to certain diseases, et cetera, and that's where a lot of the potential lies.
Chris - We know that cancer, killing one person in three, is a genetic disease. Does this mean that different cancers are going to behave differently in different tissues or that the genetics of cancer is going to differ between tissues because of what you found?
Joseph - This is one of these promising directions that we're going to do follow up research - dedicated research. So, instead of just looking at say, different tissues, we're going to look at different diseases and disease-versus-normal or subtypes of diseases, and as you've mentioned, different types of cancer et cetera, and concentrate on this. In the study already published, we concentrated on certain neurological diseases and show the relation between the code that we found and mutations in certain areas. So there's a lot of potential there definitely.
Chris - And just to finish off, you've done this in mice. Is what goes for a mouse, what goes for a person? Do the same messages hidden in the genetic sequence that make the cells chop genes up this way in mice also work in men?
Joseph - Right. So that's an excellent question. So first off, in the original paper, what we did when we analysed diseases, we analysed areas we know are conserved and we were able to relate the changes that we found using the mouse genome to diseases in the human. But of course, the next step is of course to analyse more data coming from the human. That's what actually we're doing now. So, this is sort of work in progress.










Comments
Add a comment