Phage diversity: a cast of thousands...
Interview with
Viruses are parasites that need to hijack cells to replicate themselves. We've always 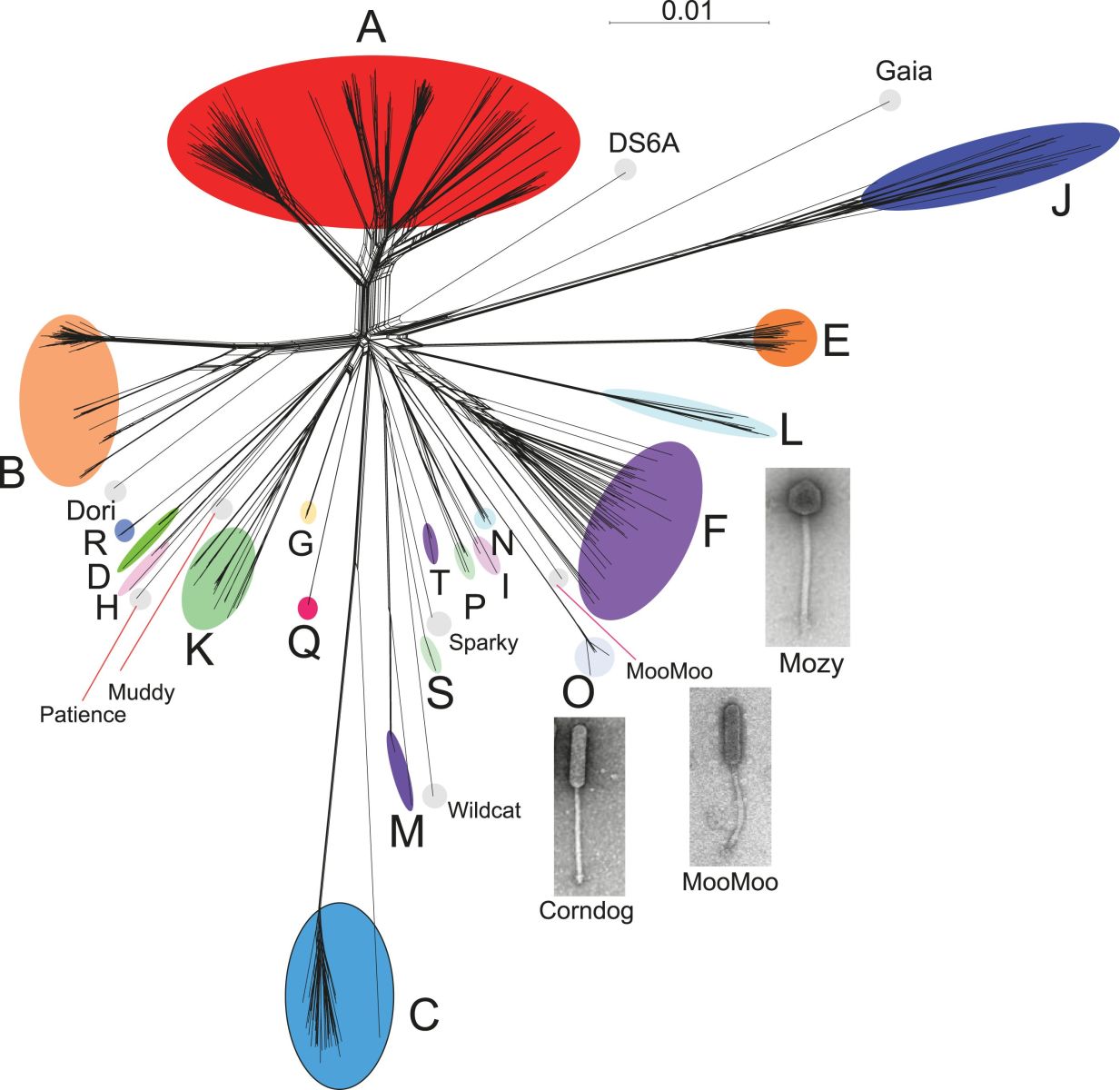 suspected that in doing so, they've probably also played a key role over evolutionary time in helping to shuttle genetic information both between cells and amongst themselves. Now a study of a group of viruses called bacteriophages, which target bacteria, shows that this is almost certainly true, as Graham Hatfull explains.
suspected that in doing so, they've probably also played a key role over evolutionary time in helping to shuttle genetic information both between cells and amongst themselves. Now a study of a group of viruses called bacteriophages, which target bacteria, shows that this is almost certainly true, as Graham Hatfull explains.
Graham - Bacteriophages which are viruses that infect bacteria, are incredibly numerous in the environment. They outnumber all other forms of life together. So, the question we wanted to address is: what is the genetic diversity of all of that information out there?
Chris - Where were you looking, specifically?
Graham - It's known that bacteriophages are very specific for their hosts, and so we chose one particular bacterial host, Mycobacterium smegmatis, and just tried to isolate the large number of bacteriophages to sequence their genomes and then compare them. And so the viruses themselves are recovered from environmental samples like soil, compost.
Chris - How do you actually extract the phages? Because I can understand if you were to go to the environment sample and then try and expand them in the laboratory, you might end up with a bottleneck there because you only select for some of them that will grow in your lab dish.
Graham - Yeah. It's a great question. And yes, it's possible that there are some phages out there that we'll miss but at least we're comforted by, in this case, using a large number of samples that have been extracted from essentially right across the United States.
Chris - To build a sort of giant family tree for this particular group of phages?
Graham - Yes. In fact, the interesting thing is that if you try to build family trees out of bacteriophage genomes, you run into an interesting conundrum. And what we find from doing the comparison is that these genomes are constructed mosaically. So, they're put together in pieces just like the Romans would build their beautiful artwork by using mosaic tiles - little pieces of stone with different colours. These phage genomes are built of segments, each segment sometimes being a single gene. Each of which has a different evolutionary history. That's an interesting finding and it means that you have to be careful about reconstructing a phylogeny, a strict phylogeny, based upon the whole genome. Because in fact what you have is an assemblage of maybe 50 or a hundred separate phylogenies which describe the evolutionary history of all the segments of that genome.
Chris - Is this arrived at through multiple infections of a single bacterial cell by multiple phages which then just basically do pick and mix for genetic segments between themselves, and then you arrive at whole host or a whole handful of new phages which have reassorted themselves effectively.
Graham - Right. And we certainly imagine that these events have been going on over a long period of evolutionary time. What is less clear is who all the participants are in this big orgy of recombination. It certainly could be phages recombining with each other, but it also could be phages recombining with resident phages that are sitting as part of bacterial chromosomes, or just with bacterial DNA itself. We suppose that the mechanism that gives rise to this process is actually an illegitimate recombination process that doesn't require the genes to be really similar to each other in order to be able to exchange. And although that might happen at low frequencies, it's also likely to be a very creative process, putting new pieces of DNA next to each other that wouldn't usually be next to each other.
Chris - And does this mean that just because you've looked at these bacteriophages that go for Mycobacterium smegmatis, that actually they could be stealing bits of phages from right across the world of phages and right across the world of microorganisms in general? And just their acting as almost like vectors, shuttling genes around all over the place.
Graham - Right. And this is exactly the kind of question that we're trying to get an understanding into. When we look at these large numbers of phages that we've sequenced, we see lots of different types. So, if they can all infect the same host, Mycobacterium smegmatis, how come it doesn't appear as though they've been exchanging more DNA with each other? So, the hypothesis would that different phages are essentially evolving by migrating across the microbial landscape, essentially changing the host that they want to infect on a regular basis. And as they make that pathway, that journey across the bacterial landscape, they essentially encounter new and different aspects or components of the gene pool with which they can essentially dip into and exchange genes and pick them up. And so that's a way in which phages can be constantly picking up vectoring and carrying around across the microbial landscape.
Chris - Graham Hatfull from the University of Pittsburgh. And the eLife paper that Graham was discussing is also a breakthrough in another way, as he revealed to me at the end of our conversation. And Graham, how many authors are there on this paper?
Graham - There's a total of 2,863, of which 2,644 are undergraduate students that participated in the isolation and the genomic analysis and the annotation of the genomes. I think that's a world's first for the total number of undergraduates on a paper. I believe the Higgs-Boson paper has the record, about 6,000 authors, but I think we're not far behind and it's mighty fine company to be in the midst of.
Chris - It certainly is and congratulations to the team.










Comments
Add a comment