eLife episode 28: From antibiotic resistance to artificial fingertips
In this episode of the eLife podcast we hear about aging, artificial fingertips, ancient DNA, antibiotic resistance and dengue fever.
In this episode
00:35 - Gender divide: why females outlive males
Gender divide: why females outlive males
with Jenny Regan, University College London
On average, males tend to live less long than females, and some diseases affect one sex more commonly than the other. So can a fly tell us why? Jenny Regan explained to Chris Smith why she thinks it can...
Jenny - In researching aging and looking at lifespan of various species, something that really sticks out is that females live longer than males. In addition to this, when we did some manipulations that could extend lifespan, there are some manipulations that extend female lifespan very well, but don’t do anything for males, specifically the ones that we were interested in is putting animals on a diet. This can really extend the lifespan of females but males derive much less benefit from being on a diet than females do.
Chris - So, how did you pursue that? What did you actually do to try and find out why there's that sex difference?
Jenny - We used the fruit fly Drosophila. What we actually started out doing was switching sex in various tissues. To do this, we took advantage of something which is particular to Drosophila biology, which is that each cell individually specifies its own sex. Whereas in mammals, you would have circulating sex hormones like oestrogen or testosterone which tells different tissues to develop to a particular gender. What you find in fruit flies is each cell individually counts how many X chromosomes it has and specifies gender that way. So, we could harness this particular feature to switch sex just in specific cell types or specific tissues around the fly. And then we subjected them to a diet and we wanted to see if having a female organ in a male fly would be able to let the males get this kind of lifespan extension that the females get in response to diet.
Chris - So what organ did you do this sex switching on?
Jenny - We looked at the liver analogue, we looked in blood, we looked in the brain, and finally, we looked at the gut. When we feminised the male gut and then put these flies on a diet, what we saw was that the males then responded to being on a diet, and their lifespan was extended.
Chris - If one studies the natural history of ageing in these flies – males versus females and you look specifically at the gut, are there clear difference between what happens in the males and then what happens in the females as they age in that organ?
Jenny - Yes, there is and this is something that we did. In parallel, we started to look at the guts of ageing males and ageing females. When we started out, we expected that we might see males had worse guts than females because males are shorter lived than females. Actually, what we found was that male guts are really well-preserved as they age. This is in real contrast to females. When we looked at females over ageing, we saw a real spectacular decline. So we saw wounds appearing in the gut and we saw small tumours appearing in the gut as well.
Chris - Can you explain why therefore on the basis of your observations, you see these difference between how long males live and how long females lives, and why calorie restriction makes a difference?
Jenny - Yes, so this is what came out of the next step of the study when we looked at female guts. Females who had been on a diet actually had better guts than females who had been fed a full complement food. And so, those respected diet females had fewer small tumours and fewer wounds in their guts. So it looked like they were better off from being on a diet. So we started to understand that perhaps this difference that people observed for the last few decades in lifespan responds to diet might be explained by the fact that the guts respond very differently i.e. the males don’t really get much of a benefit from being on a diet whereas the females do.
Chris - What is it about the female biology that means that their gut benefits in this way that the males don’t?
Jenny - The females, they're really egg machines especially towards the earlier part of their lifespan. They're laying hundreds of layers a day. So it’s really important for them to be able to get as much nutrition from food as possible. And some related studies recently have shown that females can grow their guts spectacularly when required to do so by the demands of egg production. So, it seems that females have more of a reason to have active stem cells in their gut. It also is true that females, when their guts are challenged, so by an infection, their guts respond much more than males do in the sense that they repair their guts faster or they switch on stem cells to actively divide more than males do. We think that this is probably the route of the difference that we see in the male.
Chris - So something else is holding back the males? It’s not that their gut lets them down. It’s something else that’s making them age and die prematurely compared with the females. If you sorted out the guts in the females, they would live even longer.
Jenny - Yes, absolutely. For females, the gut and the deterioration of the gut is really important. But for the males, it looks like something else is important and we think that this could be they don’t respond as well to microbial challenge so this could be something which is more of an issue for males than it is for females.
06:03 - Feeling better
Feeling better
with Calogero Oddo, University of Pisa
For people who have suffered an amputation of a hand, the current generation of 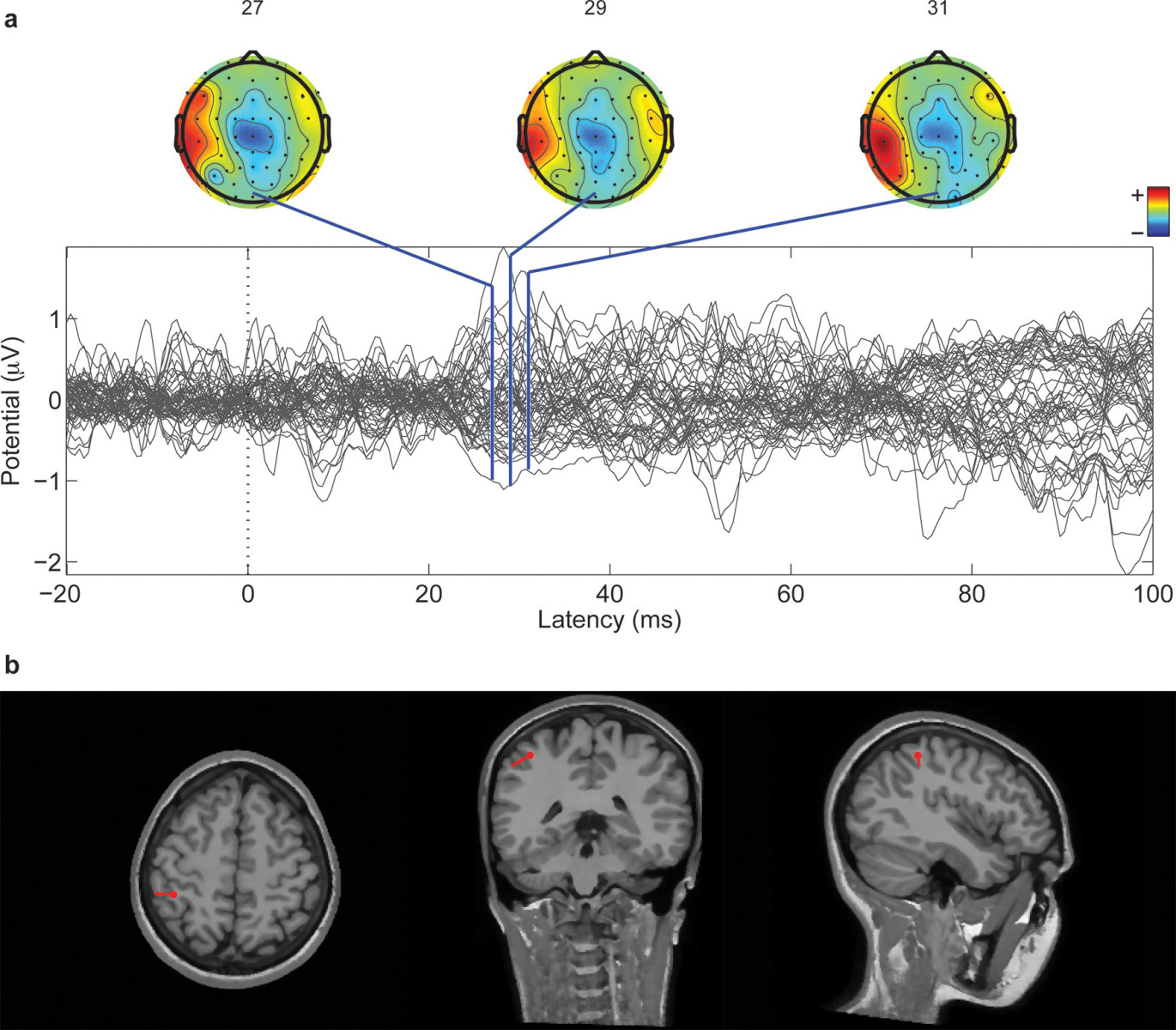 prostheses can only go so far to replacing what they've lost. They can restore some of the aesthetic and motor properties of the absent body part, but what's still missing is a sense of fine touch that can tell, for instance, whether something is rough or smooth. And this is critical for being able to manipulate objects correctly. Now we're a step closer to being able to restore this missing modality thanks to a bionic finger that uses piezoresistive sensors to detect surface textures and turn them into nerve signals that an amputee's brain can understand. Calogero Oddo is its creator, and she spoke about it to Chris Smith.....
prostheses can only go so far to replacing what they've lost. They can restore some of the aesthetic and motor properties of the absent body part, but what's still missing is a sense of fine touch that can tell, for instance, whether something is rough or smooth. And this is critical for being able to manipulate objects correctly. Now we're a step closer to being able to restore this missing modality thanks to a bionic finger that uses piezoresistive sensors to detect surface textures and turn them into nerve signals that an amputee's brain can understand. Calogero Oddo is its creator, and she spoke about it to Chris Smith.....
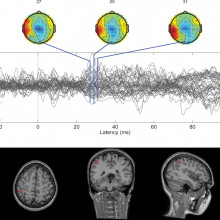
Calogero - The natural skin has the receptors internally that are the transducers. The physical interaction of our hand with the external world, they transfer mutual sequence of neural signals that are then conveyed to the nerves and then arrive through the nerves up to the brain, and then represented tactile information to our brain so that we can have a percept.
Chris - What is the material that you're working with that is capable of discerning textural features or how rough the surface is? What are you making these things from?
Calogero - The system is multilayer so we have this rubber that covers the sensors. The sensors are made of silicon which is like glass which has, inside, piezoresistors that transform mechanical information so the external information into the electrical signal. Then this is acquired by embedded electronics that converts this information into digital and then there is an artificial neuron model that keeps the continuous waveforms that are acquired from the artificial sensors. And that transforms into the spikes that are those on/off events to stimulate the nerves.
Chris - How do you teach your computer programme what each surface feels like and then how do you marry its recognition of that surface, and its output of spikes into the nervous system so that it’s putting them into the right pattern of spikes that correspond to a surface, which is what would happen in an intact individual, someone with a normal hand?
Calogero - This is done by microneurography – the technique that is minimally invasive to record from the nerves so the intact subjects when they touch naturally the surfaces. By means of this technique, we are learning how the natural code is arranged in correspondence to particular experiences and then we try to emulate.
Chris - So in essence, what you do is you're learning what the surface feels like to your particular system by recording what sorts of patterns of nerve activity a person gets when they run their natural skin over the same surface. And then when your new detectors experience that same surface contour, you just generate the same nerve output of spikes that a normal person would and that’s what you're going to put into the nervous system.
Calogero - Yes. This is exactly what we are doing. We try to capture the secrets of the natural sense of touch and to engineer it in an artificial manner so we developed a bionic finger. The bionic finger is able to reproduce to some extent the natural patterns so that are generated by the natural finger. The nice thing is that our finger is not specialised to the surfaces that we tested in this experiment. But in principle, if we test other surfaces, it is able to generalise. So, it is able to reconvert the other surface into a different sense of the spikes that the brain can interpret and recognise.
Chris - If you actually do the experiment and you take the output from your system and you apply it to the nervous system of an individual, does it really feel to a normal human that that surface is the one that they should be feeling?
Calogero - Of course, not 100 per cent but it makes sense to the brain of the subject. This was declared by Dennis, one of the amputee soldier that tested the technology. He said exactly this, not 100 per cent but it makes sense to his brain, meaning that he was able to interpret the stimulus and to imagine the stimulus in pictures.
Chris - How do you get the signals from your bionic finger back into the nervous system of the amputee in the first place?
Calogero - There was an implanted interphase that allowed the stimulus to be delivered to the nerve. What we do is insert into the nerves electrodes like wires to electrically connect the nerve and to be delivered at the stimuli.
11:15 - Why did plagues stop?
Why did plagues stop?
with Kirsten Bos, University of Tuebingen
In the early middle ages up to half of the population of Europe were wiped out by the  Black Death, caused by the bacterium Yersinia pestis. But, following its arrival, the initial pandemic wave waned and was replaced by a series of localised epidemics around Europe until the 18th Century, when it abruptly disappeared. So how do we account for this behaviour? From the University of Tuebingen, Kirsten Bos explained why to Chris Smith...
Black Death, caused by the bacterium Yersinia pestis. But, following its arrival, the initial pandemic wave waned and was replaced by a series of localised epidemics around Europe until the 18th Century, when it abruptly disappeared. So how do we account for this behaviour? From the University of Tuebingen, Kirsten Bos explained why to Chris Smith...
Kirsten - We’ve identified the strain of the bacterium responsible for bubonic plague, Yersinia pestis in archaeological material from Marseille, France from 1722. Plague was at one time a very pervasive infectious disease in Europe. We know it most famously from the Black Death. However, the disease persisted in Europe for the next several hundred years as local epidemics. However, for some reason, it’s gone extinct in Europe. We don’t have large plague infections today within the European continent. So the question becomes, why did pestis go extinct? Why did these plague epidemics stop happening. We wanted to address this question by looking at archaeological material first to type the strain that was present in some of these older European pandemics.
Chris - I suppose it boils down to: did something change in the bug, or did something change in the host whether that’s the rodent that carries it and then gives it to us, or even all of these factors – the environment, the host down to the pathogen itself?
Kirsten - Certainly. So what we’ve done up to now is we have looked at the genetic profile of the bacterium that caused the great plague of Marseille from 1722. We have found that this lineage surprisingly is an extinct lineage or so we believed it to be extinct because it’s not been typed in any modern pestis material. So with that, it appears that pestis was probably living somewhere in a population in Europe itself or easily accessible to Europe, perhaps in west Asia. And that might have been contributing some of these pandemics or epidemics that we were observing within Europe from the time of the Black Death until the mid-18th century.
Chris - What was the material from Marseille that you were analysing to get this data?
Kirsten - We used teeth from a large plague pit that was excavated in the mid-90s. Teeth are excellent material to use for this type of analysis because the thick enamel coating on the tooth actually functions like a shell to preserve the DNA that’s in the interior. In the interior of the tooth, you find an area that’s called the pulp chamber. This is highly protected archaeological material and that’s the best part in the skeleton to go if you're trying to look for a bacterium that might have been presented in the bloodstream of an individual at the time of death.
Chris - Other archaeologists, yourself included have looked at teeth from other plague sites including London from periods earlier on in the course and history of plague. How does the sequence profile that you’ve got from Marseille compare with those earlier victims?
Kirsten - Interestingly, we find that the strain we’ve typed here is a direct descendant of the strain that was circulating in London at the time of the Black Death. Even more interestingly, this strain does not show any close genetic similarity to any strains that are circulating today. To the best of our knowledge, this strain is currently extinct. So what this tells us is that this was probably a daughter population of the strains that gave rise to the Black Death. The big question is, where was that daughter population residing during that second wave, so between the 14th century and the mid- 18th century.
Chris - You think there's therefore some kind of reservoir whether that’s a human or an environmental reservoir which keeps receding populations over hundreds of years and eventually just peters out.
Kirsten - Yes, that’s correct. We figured there was a plague population somewhere. A compelling plausibility is that it was somewhere within Europe itself and is just no longer present or it could’ve been somewhere very close to Europe and accessible to it by marine transport so for example in west Asia. We can't identify where that reservoir was, because of course, with genetic data, we’re providing only one important piece of the puzzle. It’s important to know that this lineage appears to be extinct. However, where that plague reservoir was residing is going to require information from historians so we can determine where people were moving and when. We need to get information from zoologists who learned what plague species were present in different areas at different time periods. We need to get information on climatic data because there's a relationship between climate and bubonic plague outbreaks to arrive at some hypothesis about where plague might have been residing in that very essential time period.

16:00 - Antibiotic resistance genes in the meat industry
Antibiotic resistance genes in the meat industry
with Noelle Noyes, Colorado State University
Antibiotic resistance has been highlighted as a major medical risk facing the global population. And as bugs become increasingly difficult to treat, antimicrobial drug pipelines are ironically almost empty. One source of drug resistance might be the meat industry where antibiotics are administered to animals to keep them healthy and to encourage them to grow rapidly. But what risk does this pose? Colorado State University's Noelle Noyes has been following the food chain, as she explained to Chris Smith...
Noelle - The meat that we eat could potentially harbour different organisms or different antimicrobial resistance genes that we could ingest and then could make us ill. But other way that this can happen is the fact that we’re all connected environmentally in some way or another, and to some degree or another. So, these animal production facilities are part of our environment and things that happen in these different animal production facilities can be dispersed into the environment through different pathways.
Chris - How then were you trying to find out what the likely traffic is for these antibiotic resistant elements between the agricultural setting and the human world?
Noelle - The thing that we would love to do, the ideal study would be to take a group of animals and to track them throughout their entire lives, all the way from the time they're born on a farm until they reach your dinner plate. That’s incredibly difficult to do given the livestock production system. So in this study, we tracked pens of cattle from the time they enter the feedlot through the time they exit at the feedlot then their transport through the slaughterhouse. And then we actually followed the animals into the slaughterhouse and then the carcass was disassembled and we actually sampled the final product right before it was packaged for distribution into the retail chain.
Chris - What source of specimens were you collecting at each stage and how did you analyse those specimens?
Noelle - We collected faecal samples from the pen floor, soil from the pen floor, and then we collected drinking water samples. When they were unloaded at the slaughterhouse, we went into the trucks and we swabbed the inside walls of the truck. The cattle then go through the slaughter process and at the end of that, we swabbed the conveyor belt.
Chris - How did you analyse these things? were you trying to culture microorganisms? Were you using genetic techniques to look what was in there?
Noelle - We don’t use culture and we don’t use PCR. We basically extract all of the DNA that are in all those samples that I just described and we fragment that DNA into small pieces and then we sequence it.
Chris - And you can then match that up with a database so you know what bugs are there and potentially, what antibiotics they have the ability to fend off.
Noelle - Yes. So we take those short fragments of DNA and we basically just try to match the pattern to known resistance gene DNA sequences that are available publicly.
Chris - What did this reveal to you when you did it? What did you find?
Noelle - The first finding that surprised us initially was that when we took those samples at the very end of that production system and we did this method, we actually didn’t identify any resistance genes. So that was a surprising finding. And then the other finding was what was happening during the feedlot period. So that’s when antibiotics tend to be used more frequently in cattle production. During that time, we found a decrease in the diversity of the different resistance genes. The genes that tended to persist during the feeding period were resistance genes to antibiotics that were used in the cattle so that made sense to us.
Chris - That reflects a sort of selection within the animals for bugs that are endowed with that resistance so they can enrich in those animals because you're giving those animals those antibiotics.
Noelle - Yes. So, that’s the theory. Of course, a lot more work needs to be done to figure out if that is really what's happening. But the hypothesis at this point is that the antibiotics that were being used were creating a selection pressure that through natural selection favoured the bacteria that were able to defend against those antibiotics. And so, we saw those resistance genes persisting in the bacterial population throughout the feeding period.
Chris - Late in 2015, there was a study. It was published in one of the Lancet journals showing that in China, there was a significant co-occurrence of resistance against one class of antibiotic. This was colistin where large amounts of it were being used and it was cropping in the meat but also patients in hospitals. So how does that discovery sit alongside your work?
Noelle - Yeah. It speaks to the same point which we talk about in the paper which is that we really need to perhaps not shift focus but maybe expand focus to looking at these potential environmental routes of transfer. So, we have very robust systems right now, surveillance and monitoring systems, as well as food safety systems that sort of deal with that food chain transmission route. But we have much less clarity around the environmental exposure route. And so, I think that that paper that you just described as well as our work suggests that perhaps we should expand our focus to include some of these potential environmental pathways.
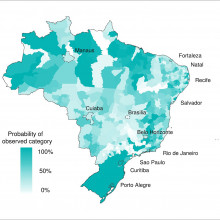
21:12 - Predicting Dengue Outbreaks
Predicting Dengue Outbreaks
with Rachel Lowe, Catalan Institute of Climate Sciences in Barcelona
Millions of people succumb to dengue virus infection each year. It's spread by the Aedes family of mosquitoes and causes a haemorrhagic fever, which can range in severity from mild to lethal. But predicting where the hotspots will be, so that appropriate healthcare provisions and anti-mosquito measures can be supplied to the right places is very difficult. Rachel Lowe, from the Catalan Institute of Climate Sciences in Barcelona, has been tackling the problem with a bit of help from FIFA, as she explained to Chris Smith...
Rachel - We developed a prototype dengue early warning system to produce probabilistic forecast of dengue risk 3 months in advance. We specifically designed this system to predict the dengue risk ahead of the 2014 World Cup in Brazil. So, which areas of the country were at a greater risk of experiencing epidemic levels of dengue.
Chris - So, you had the prediction and then of course, now we’re 2 years on from the World Cup, you can test whether or not your model accurately predicted what was going to happen.
Rachel - Yes. We have now compared the observed dengue incidents rate during the World Cup to our predictions to see in which areas of the country the model worked better and was able to successfully predict high levels of dengue risk.
Chris - And was it any good?
Rachel - We found that in the areas experiencing high levels of risk, the model successfully predicted 57 per cent of those areas. We also compared this to a model just based on the historical seasonal averages of dengue. We found that our model was considering better.
Chris - Some people would say well, 50 per cent-ish, that’s not particularly good. Why is it not 100 per cent?
Rachel - Of the areas experiencing high risk, the forecast model was able to predict 57 per cent of those areas compared to 33 per cent just based on seasonal averages. So, current practices is based on generally knowledge from the past to be able to support decisions. So we found that by incorporating seasonal climate information along with epidemiological information 3 to 4 months in advance, we have this added 24 per cent of areas we’re detecting high risk. Of course, because we’re using quite a long lead forecast several months in advance, it’s very difficult to accurately predict 100 per cent of these areas which depends not only on the climate and early cases of the diseases but also on many other factors such as the types of the virus that are circulating and the susceptibility of human populations and how they're behaving on the ground.
Chris - So actually, it is significantly better than what would be achieved if one just said, “Well, where does it normally produce a hotspot? What's been the seasonal average previously?” what do you attribute the improvement to? What's the deciding factor here?
Rachel - Well, this model is able to take into account variations in the climate in space and time. so by incorporating that information which changes from year to year then you can have a better idea of how the mosquitoes might respond to these particular climate characteristics. And also, by incorporating early cases of dengue from the surveillance system several months in advance, you have some sort of indication of what might happen in the coming season.
Chris - More than a hundred countries worldwide have now got dengue activity and it’s a major disease burden internationally so could I take what you’ve pioneered here in Brazil and apply that to say, Indonesia or the top of northern parts of Australia where we do see dengue activity and use the same parameters to make equivalently good predictions there?
Rachel - You could certainly use the same model framework and it would be wise to take data from those areas and formulate the model based on the dengue incidents and the climate conditions, and other socioeconomic factors particular to that location. But we have certainly developed this model framework for other locations such as Thailand and Ecuador.
Chris - Now obviously, the one thing people are very, very worried about at the moment in the same country Brazil is zika virus because they spread in very similar ways. Could we take this model and also now make predictions about other viruses entirely like zika?
Rachel - Absolutely. this model is a climate base model and many diseases including dengue, chikungunya and zika which are transmitted by the Aedes mosquito are sensitive to climate. So this model could certainly be extended to try and predict the risk of the transmission of other viruses such as chikungunya and zika.










Comments
Add a comment