How To Tell Coronavirus Variants Apart
We’re looking at the most critical variants of the coronavirus, and finding out how to tell whether they’re flummoxing COVID vaccines. Plus: the cost of catching a serial killer; DNA with four strands instead of two; and a mutant fish whose fins have started turning into limbs!
In this episode

00:33 - Coronavirus variants: the name game
Coronavirus variants: the name game
Sharon Peacock, COG-UK Consortium
You might know them as the "UK variant" or "South African variant" - but what are they really called, and why? Phil Sansom has your coronavirus variants field guide, with help from head of the COVID-19 Genomics UK Consortium, Sharon Peacock...
Phil - To an extent, any talk of new coronavirus variants is slightly misleading.
Sharon - Looking for new variants is like looking for a needle in a haystack because there are thousands and thousands of new mutations in the virus. And so looking for one that's important can be very challenging.
Phil - That’s Sharon Peacock, talking to Chris Smith on the Naked Scientists in January. She runs a consortium called COVID-19 Genomics UK, or COG-UK. They feverishly sequence the full genetic code - the whole genome - of the samples of coronavirus they get sent, and so far have more than 200,000 sequences under their belt.
Phil - The point she’s making is that it’s easy to find a new variant - it’s much trickier to find a variant that’s both new and more dangerous. There are a few methods.
Sharon - First of all, we can look at spread between populations. We can also look at the population of genetic information that we have to demonstrate whether, for example, a particular lineage or line is expanding more rapidly than we expected. But laboratory experiments are really critical here, they provide crucial evidence, and there's different types of laboratory experiments that we would want to do. First of all, we'd look at how the virus interacts with cells that are grown in the laboratory: are they more likely to adhere to those cells and get entry into them? We'd also want to see how the virus interacts with antibodies that have been raised by people who've had a natural infection or been vaccinated, to see whether that interaction is what we would expect or not.
Phil - All of these techniques became very useful in the UK at the end of last year, when they discovered what later came to be known as the UK variant, the Kent variant, or B.1.1.7.
Sharon - It was only really in mid to late November when it really became clear that there seemed to be spread of cases in the South of England, whereas there wasn't spread in other parts of the country that were under similar cut a lockdown rules. There are range of explanations for that, but one of them is that the virus could be somewhat different in its behaviour. And so at that point, in the beginning of December, analysts who understand the genomes very well realised that the two could be related: an increasingly transmissible virus linked to a very specific genome.
Phil - B.1.1.7’s genome is specific by 23 mutations, and it’s also not clear where exactly it came from - it wasn’t particularly related to other variants going round the UK. That was in December. A few days later, South Africa announced they’d found their own concerning variant. Then within the space of a month, two were found in Brazil. Turns out pandemic gut punches sometimes come like buses. But Sharon would question the way we’ve been referring to them.
Sharon - I think it's really important that we avoid labelling variants based on where they've been first detected, because where they've been first detected doesn't necessarily mean where they've emerged. And nobody can really be held responsible for an act of nature - for a mutation.
Phil - Sharon Peacock, head of COG-UK. So bearing that in mind - can we decode the names these variants have got? They tend to be complicated because genetics is complicated, and the WHO hasn’t yet come up with a standard naming system. Into this void, a few main systems have emerged. The open access-database GISAID uses one system; the group Nextstrain that we featured on Naked Genetics back in April uses a second.
The third is my favourite, and it’s where the name B.1.1.7 comes from. It’s a system proposed last year with the acronym PANGOLIN. Here, instead of defining a variant by what’s in it, you define it based on events in its past. Those events will usually be: it arriving in a new place, and expanding and multiplying through the area.
Using their website, we can explain why it’s called B.1.1.7. “B” was an escape from China at the end of 2019; the first "1" is a big expansion through Europe at the time of the Italian outbreak at the start of 2020. The second “1” is a more specific European wave, and the “7” is the bad news variant we’re dealing with now.
Let’s translate the other key variants now. The South African variant in PANGOLIN is B.1.351. The Brazilian variants are called P.1 and P.2, which are shortened versions of longer names in the B.1 group.
These names are clearly many times less memorable than naming them by countries. We’ll just have to do our best to repeat them until we remember: B.1.1.7, B.1.351, and P.1 and P.2.
05:20 - Pfizer vaccine: can it handle new variants?
Pfizer vaccine: can it handle new variants?
How concerned should we be about new coronavirus variants? The evidence is still coming in. They all seem to spread faster, and there are hints that a couple may possibly be more lethal, or make serious illness more common. The real question, though, is whether vaccines still work against them. That’s what University of Texas biologist Pei-Yong Shi is trying to answer - as he told Phil Sansom...
Pei-Yong - Based on all the data so far we've accumulated, we think that, at this point, the vaccine still works quite well against the mutant viruses we've tested so far.
Phil - That's a relief...
Pei-Yong - Yes. Using what we call the reverse genetic system, you can make the virus, and then manipulate it at any position you want; and then through animal models and the cells, we can really look at their properties.
Phil - Pei-Yong’s team pioneered this “reverse genetic system”, a method for rapidly generating viruses with exactly the mutations you want to study. They’ve partnered with Pfizer and are receiving samples of blood from people who got the Pfizer vaccine. They then test the blood on their engineered viruses to see if the immune response still works. The first thing they’re comparing is individual mutations rather than the combinations of them you get in these variants.
Pei-Yong - There are prominent mutations such as the 501Y mutation. And that mutation is very particular because it's not only recovered from the UK variants, but also it was recovered from the South African strain and the Brazilian strain. And what we found is that mutation doesn't seem to affect the inhibition of the viruses by the immunised blood.
Phil - Basically no difference with this new mutation.
Pei-Yong - Yeah, very, very minor differences, yeah. If there is any.
Phil - That’s a relief, because N501Y is one of two crucial mutations found in the bit of the coronavirus that binds to human cells. The name is referring to amino acids, which are the building blocks of proteins that the genes are coding for. Here, thanks to the mutation, amino acid number 501 has changed from a type called N to a type called Y. Pei-Yong has also tested the second of the two crucial mutations.
Pei-Yong - In the South Africa strain there is this mutation '484'. And this change has been well documented before: that once you have this change, the virus will reduce its sensitivity to monoclonal antibodies. And that is a very worrisome mutation. In order to study that, we did a similar kind of trick. We do see it very modestly reduces the inhibition activities of the blood; but this reduction decrease is very minor and modest.
Phil - Right, so in combination, how much are they reducing the vaccine's power by?
Pei-Yong - In the South African strain, you can consistently see there are seven mutations or deletions. Just now we're just talking about one by one. The question is, if you put them all together, what is the total effect? That is still ongoing; maybe next week we'll get all the answers to that. Which what we did is: we made a virus that has all those seven mutations, and then we are now testing how that is going to affect the total inhibition by the vaccinated blood.
Phil - Wow, this is a quite a big job, isn't it? Because there's a bunch of variants out now. Do you have to test all of them?
Pei-Yong - Well, that's what keeps us very, very... whether they add together or they are counteractive... this the only way we'll be able to get to the bottom of these functions.
Phil - And then do you have to test each version against each type of vaccine as well?
Pei-Yong - I would think the results can be generalised to the different vaccine platforms, because they're all encoding the same sequence of the spike protein. But at the same time, we should be very cautious.
Phil - And then what's the point where you see something that makes you go, "oh boy, time for a new vaccine"?
Pei-Yong - Before we can reach that consensus or decision, there are still a lot of questions we don't know. For example, what is the minimum antibody level that is required to protect us from being infected? That minimum bar, that number, has not been defined at all. And this is a very, very important number. Let's say after people get vaccinated by Pfizer's vaccine, they have an average antibody level of '600'. Now let's say with the new variants, 600 becomes 200. 200, I think, is still way above the minimum requirement to get us protected. So there is a misconception: if the antibody level is reduced by threefold, that doesn't mean the efficacy is going to drop by threefold. But unfortunately right now don't know what is the minimum bar required.
Since recording Pei-Yong Shi has reported in the New England Journal of Medicine the work testing the full combination of mutations in the crucial part of variant B.1.351, the variant from South Africa. Pfizer’s vaccine works only a third as well against it compared to other variants.

12:38 - Golden State Killer caught using family tree sites
Golden State Killer caught using family tree sites
Libby Copeland, Journalist
In 2018, US police found and arrested a serial killer in a brand new way. They connected his DNA to the DNA of a relative who’d uploaded theirs online for genealogy. For many, the ends here didn’t justify the means; it felt like a serious breach of privacy. And at the end of 2020, the Los Angeles Times reported that the breach was much wider than anyone had realised. Independent journalist Libby Copeland told Phil Sansom the story...
Libby - The origins of the Golden State Killer case from two and a half years ago were not quite what we understood them to be. They involved more databases and private databases in a way that Americans and the rest of the world had not previously known.
Phil - Right, you better give me some context: who's the Golden State Killer?
Libby - His name is Joseph James DeAngelo, and during the seventies and eighties in California he went on a reign of terror, raping and murdering... obviously many, many people. He was also briefly a police officer. He was somebody who was never suspected, never implicated; and when he was arrested in 2018, what came out is that instead of using all the traditional methods that police typically use, they had actually used consumer DNA. People willingly swab or spit into a vial, they send it into a company, and they get back those pie charts and those lists of DNA relatives. And the breakthrough in the Golden State Killer was to take those techniques and to apply them to crime scene DNA that had been left behind.
Phil - Let me get this straight: this guy, and everyone else, basically thought that he'd gotten away with his mass murders for decades, until what - a relative uploaded a bit of DNA and they caught him with that?
Libby - Yeah, it was apparently somebody who was closer than a third cousin, but quite possibly somebody who didn't know him.
Phil - Is this kind of a first?
Libby - It's a big deal. When investigative genetic genealogy - which is this new crime solving technique - emerged back in 2018 it was a really big deal, but it was limited in its scope. We understood that it affected only one database: a kind of obscure hobbyist database that was publicly accessible, that was volunteer run, and that was free. So the stakes were not so high. What the Los Angeles times reporting revealed is that, in fact, three databases were involved; and in addition to GEDmatch, that free, publicly accessible site, one of them is called FamilyTreeDNA, and one of them is the third largest database for ancestral DNA. It's called MyHeritage. What's really striking about this is that MyHeritage did not know.
Phil - Aren't these databases designed to help people connect? And if so what's the big deal? The police just did something that everyone else is doing.
Libby - Right. That's a very good point. And that's been the response from a lot of people who are in support of this, and indeed from the civilian genetic genealogist who did the upload. Her argument is that this kind of use of DNA to find genetic kin is something that has been happening, at this point, for over a decade. If you think about the anonymity that sperm donors were promised, for instance: those are men who, say, back in the 1970s they contributed their sperm to help other people start families, in exchange for pay. And they were often contractually promised anonymity. And DNA has done away with that, right? It's made those contracts moot. There's no such thing as an anonymous sperm donor anymore. So her argument is: if anything, sperm donors are more entitled to privacy than, For instance, a rapist who left his DNA at the scene in the act of committing a rape. On the other side, of course, individuals have different - and less - power than law enforcement does. Law enforcement has the power to arrest. And the question is: in the context of DNA, is law enforcement entitled to go wherever the public goes?
Phil - When it comes to the police and law enforcement, it always feels like the things that we know about are the tip of the iceberg. What do you think is going on that we don't know about?
Libby - I think it is certainly possible that this has happened in other cases in addition to this. And I have heard people who are knowledgeable say that they believe that this has happened before.
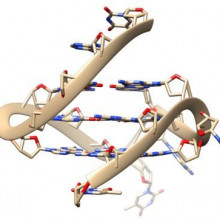
17:15 - G-quadruplex DNA found in human cells
G-quadruplex DNA found in human cells
Ben Lewis, Imperial College London
What shape is DNA? The helter-skelter double spiral you’re thinking of is mostly right. But sometimes DNA tangles itself into stranger shapes - and now scientists from Imperial College London have found evidence for one of these inside human cells. It’s called the G-quadruplex, and researcher Ben Lewis explained it to Phil Sansom...
Ben - You might know about the double helix. And if you know anything about it, you might know how it was one of the most pivotal discoveries in all of genetics. The story didn't end there, because DNA can actually adopt other structures. A lot of these are very unusual and wacky, but we're interested in one called a G-quadruplex; and the reason we're particularly interested in this is because it's stable and can form in the same conditions as inside our cells.
Phil - What is a G-quadruplex?
Ben - It's a quadruplex as opposed to the normal structure of DNA, which is a duplex. So duplex means we have two strands; and in the G-quadruplex instead we have four strands, the DNA wrapping back on itself four times, to form this structure which is often seen as being like a knot.
Phil - How strange!
Ben - It's certainly unusual, but there's all kinds of amazing DNA structures that people are interested in. Perhaps it's not the most surprising that one of those can actually form in the same conditions in ourselves.
Phil - How can you tell? DNA is very small!
Ben - Yes, so being able to see G-quadruplexes is the challenge. There's going to be so few of them compared to the rest of the DNA, and it obviously exists amongst a huge amount of other DNA; so we're trying to find a needle in a haystack, except that needle is also made of hay.
Phil - Then how do you do it?
Ben - So the normal way that you would try and find something inside a cell is by creating a probe which binds specifically to that thing you're looking for, and then gives out some signal that you can see. Most usefully you would do something fluorescent. But the problem with that is that you need a probe that's super specific, and there's just so little G-quadruplex DNA, it's really almost impossible to make a probe that's specific enough. So we've had to use a slightly more complicated approach to try and overcome this hurdle.
Their approach uses a fluorescent probe like Ben describes, but it measures the light in a different way.
Ben - Normally you would look at all the light that comes from one point. But instead we look at when it arrives at our detector, and this is called the fluorescence lifetime. The fluorescence lifetime is affected by the environment around our molecule. So we don't need a super specific probe for our G-quadruplex; we just need a probe that gives us a different florescence lifetime when it's bound to quadruplex DNA compared to all the other DNA. And that's what we've been able to make a probe that's able to do.
Phil - Right. So now that you can figure out where the G-quadruplexes are - when you look at human cells, what do you find?
Ben - There are quite a number of G-quadruplexes present in the sales we've studied. The way we've done this is by looking at the machinery that people think is handling and unwinding these G-quadruplexes in ourselves. And for the first time, we've actually been able to see that when you get rid of this machinery, you see a lot more of the G-quadruplex DNA inside those cells. As of right now, we don't really know exactly how many G-quadruplexes there are; but now that we have a new tool to use which allows us to look for these G-quadruplexes in living cells, we have the opportunity to start answering questions like that, and many others.
Phil - Another question then is: if you've got special machinery inside you that gets rid of this stuff, is it something that's only there by mistake, and it's a problem?
Ben - Well, our cells have a great knack for using all the tools available to it. So whilst you could see in some circumstances these knot-like structures getting in the way of our normal cells' processes, there are also a lot of potential ways in which the cell might use these quadruplex structures as markers that tell it when to potentially express specific genes, or for other uses that we haven't even imagined yet.
Phil - Like for what? Can you give me an example?
Ben - These G-quadruplexes could be used or abused in the case of cancer cells. There's a lot of sequences of DNA that could form G-quadruplexes just before genes that are vital to cancer cells, where the cells hijack these genes and use them to grow out of control into tumours. But at the same time, we've also seen ways in which the cells could use these G-quadruplexes to stop cancer in its tracks. This is particularly seen in the ends of our chromosomes, in a part called the telomeres; these are the protective caps that protect the end of the chromosome from damage, in the same way that the little plastic caps on your shoelace protect the ends from fraying. There's a lot of sequences there that could form G-quadruplexes, and these have been shown to actually be able to stop the machinery that cancer cells use to extend those caps indefinitely and help them become immortal. The formation of quadruplexes could actually put a halt to that process and prevent those cancer cells from successfully immortalising themselves.
23:04 - The mutant fish whose fin is becoming a limb
The mutant fish whose fin is becoming a limb
Brent Hawkins, Harvard Medical School
When our ancestors first crawled out of the oceans, among other things, they needed to evolve limbs to get around. And now, millions of years later, scientists from Harvard have helped some fish repeat the process - by finding a mutation that makes their fins seemingly start to transition into arms. Does something smell fishy? Phil Sansom went to study author Brent Hawkins to investigate…
Brent - What I found with my colleagues is the unexpected ability of a fish to transform its fin into a more limb-like configuration. And we found this in mutant zebrafish; zebrafish are kind of like the lab mouse of the fish world.
Phil - So when you say unexpected ability - this isn't one fish that can do it as a superpower?
Brent - No, no. It's actually a whole line of mutant fish that reliably reproduce this change in their fins across generations.
Phil - You said zebrafish: what is a zebrafish?
Brent - Zebrafish are small... what we call teleost fishes, that's sort of the family they're part of. They're originally from India. And back in the eighties and nineties, when geneticists were trying to find a nice genetic model that they could do these very large genetics screens in, the zebrafish was the recommended species to try, because people knew in the aquarium you could mate them very easily and get lots of eggs. You basically just need to put a male fish and a female fish together in a little tank overnight, and in the morning when the lights turn, that cues them to spawn. So it's very, very convenient.
Phil - How were you then mutating them?
Brent - The way we do mutations in the fish is we take a male fish and inject it with a chemical called ENU that's going to cause random mutations in its sperm; made it with a female; and then we look at their offspring, and those offspring will have inherited the mutated genes from the father.
Phil - You're sort of throwing genetic spaghetti at a wall then, to see what sticks and looks interesting?
Brent - Exactly. That's perfect.
Phil - So what was special about this one then?
Brent - A normal zebrafish has these very nice stripes along the body, but our mutant has a more scrambled pattern. And so that's why we picked it out initially. But in our screening process, we look at many different parts of the fish, and one of the last things we look at is doing our processing to look at the skeleton. And it was at that step that we saw that the pectoral fin had these new bones that shouldn't be there.
Phil - Was it a moment of just sudden realisation?
Brent - Oh yeah, it was a total dumbfounded Eureka moment when we saw it! It's something that shouldn't be there, ever. A normal pectoral fin has a very reduced skeleton compared to our limbs: just these series of four short bones arrayed side by side. What happens in the mutant is that instead of just having these four side by side bones, they make two more bones further away from the body, on top of those four side by side bones.
Phil - Are they sort of jointed on to the old bones?
Brent - Yes they're well-integrated! They have joints and they have musculature that allow them to articulate with the other bones.
Phil - That's a huge change!
Brent - Oh yeah. It was very surprising that with one single mutation, we got this coordinated shift in not just the skeletal system, but also the muscular system and the formation of the joints as well. And through our mapping, we did determine it is one single gene causing all of these changes.
Phil - What gene?
Brent - The mutation is residing in a gene called waslb. The waslb gene is involved in modifying microfilaments in the cell, that are involved in all sorts of processes like cell migration and displaying receptors. The gene is also involved in the transcription of other genes.
Phil - One of those pathways must be sort of a finger bone type pathway, right?
Brent - That's what we're hoping. And that's what we're looking at now more carefully. So for instance the hox genes, which have important roles in patterning many different parts of the skeleton; we found that these hox genes are actually in part controlled by the waslb gene. We know hox is important for the limb, so this is an exciting new, additional level of regulation that we didn't know about before.
Phil - Do you know then, Brent, whether you've recreated a step that some of these fish's ancestors way back would have taken to help get out of the water?
Brent - It's possible. In our research, we're only looking at the living fish; we haven't recreated evolution by any means. But what we do show is this hidden ability in a fin to become more limblike.
Phil - I just can't believe you're getting all this from a single mutation. Imagine what you could get with another two or three!
Brent - Oh yes, the prospects are wonderful! And that's actually something we're doing now: we have other genes that cause similar changes in the skeleton, and now we want to put those together in the same fish. And we're really excited about that - that one day we might have fish crawling out of the tank.
Related Content
- Previous Restoring Old Masterpieces
- Next Learning In The Time Of COVID










Comments
Add a comment