What is the scale of the superbug problem? How much is antibiotic resistance costing? Can new antibiotics be made that cannot be bypassed by bacteria? And what new drugs are already in the pipeline. In this infectious episode of the Naked Scientists, we put the rise of antimicrobial resistance under the microscope and ask what scientists are doing to combat the problem. Plus, why the abominable snowman hasn't been discovered... yeti, 46-million-year-old blood from a fossilised mosquito, phage therapy for C. diff and the brain wash-out that happens when we sleep...
In this episode

01:26 - Could the yeti be a bear?
Could the yeti be a bear?
Much to the disappointment of cryptozoology fans everywhere - that's the study of 'hidden' animals such as Bigfoot, or the Loch Ness Monster -  a scientist from Oxford University is claiming that the legendary ape-like Himalayan monster, the yeti, may be nothing more than a type of bear.
a scientist from Oxford University is claiming that the legendary ape-like Himalayan monster, the yeti, may be nothing more than a type of bear.
Professor Bryan Sykes has analysed hair samples from the mummified remains of a yeti-like creature shot by a hunter in the mountains four decades ago, and a single hair that was found ten years ago in a forest in Bhutan by a group of yeti-hunting film-makers.
He compared DNA from these samples with sequences from other animals in the international DNA database and found a 100 per cent match with a jawbone from an ancient polar bear found in Norway, dating back tens of thousands of years. This is around the time that polar bears and brown bears are thought to have separated into two species, although they're very closely related and can interbreed where their territories overlap.
Sykes thinks that the most likely explanation for the yeti myth is that the animals are actually some kind of brown bear/polar bear hybrid rather than an ape. But as nobody has seen a yeti alive or conclusively got a body, it's hard to know for sure. And it doesn't mean that there are rogue ancient polar bears roaming the mountains today - he thinks it's more likely that bears (if they exist) mistaken for yetis today are hybrids.
This isn't the first time that analysis of alleged yeti samples have failed to come up with evidence for that it's an ape-like species - in 2008, a similar analysis on hairs from the north Indian Himalayas turned out to be from a type of mountain goat, rather than the ape-related species that many cryptozoologists believe Bigfoot to be.
This research also comes with a bit of a scientific 'health warning' as the results haven't yet been published in a peer-reviewed journal, so other scientists haven't been able to see the data and comment on them. But once that happens, we'll know more about the likelihood of the bear theory being correct...
04:23 - Fossilised mosquito meal
Fossilised mosquito meal
Blood from the last meal consumed by a mosquito 46 million years ago has been identified inside a  fossilised mosquito found in Montana.
fossilised mosquito found in Montana.
Writing in PNAS, Dale Greenwalt and his colleagues at Washington DC's Carnegie Institution, initially used X-rays to examine the remains of a female mosquito preserved in a layer of fossilised mud that would have rested on the floor of a pond in what is now north-western Montana.
Consistent with the mosquito's engorged appearance, the X-rays showed a strong signal corresponding to iron, which would be in keeping with the insect having binged on a blood meal not long before it presumably drowned in the pond.
Next, the team cleaned away the surface of the fossil to remove overlying material, and then used a technique called "time of flight secondary ion mass spectrometry" - ToF-SIMS - to probe the chemical composition of the insect itself.
In this system, charged particles are fired at a surface where they knock out molecules that can be sucked up and identified by the analyser.
Stable molecules called porphyrin rings, of which haemoglobin is one, were detected, giving a signature closely matching pig blood tested as a control.
This suggests that blood has been preserved within then fossil, and that some of the chemicals sucked up 46 million years ago by the now-fossilised female mosquito are still preserved within its remains today.
Ironically, this paper, which comes 20 years after the Hollywood blockbuster Jurassic Park in which dinosaur DNA was recovered from mosquitoes preserved in amber, is actually the first such description of a blood-engorged fossilised mosquito!

07:12 - Invasive Species
Invasive Species
It's official: the UK has a slug problem. This week, researchers from the John Innes Centre in Norwich asked the public for help to help them track down the Spanish slug, a rapidly reproducing invasive species that eats crops and is not deterred by slug pellets. Here's your quick fire science on invasive species, with Matt Burnett and Simon Bishop...
- An invasive species is any animal or plant that has come from another area, which disrupts the environment and outcompetes native species.
- Humans like to travel the world, and sometimes other species like to come with us. Mosquitoes spread to some Pacific islands by hitching a ride in the wooden planks of boats. Unfortunately, they also took Dengue fever with them.
- Dutch Elm disease has also been ravaging woodlands in Europe and North America after accidental introduction from Asia. Our trees just weren't prepared for it, having never needed to evolve resistance.
- Zebra mussels from Russia spread around the world in ships. It costs the UK water treatment industry millions of pounds every year to clear them out from pipes.
- Species have also been introduced to countries just because they are exotic. Japanese knotweed was brought to Britain in the 19th century because people thought it looked pretty, but gardeners soon found that it can grow through concrete, damaging everything from roads to drainage systems.
- The American crayfish was brought to Britain to give farmers extra income, but it escaped the farms and outcompeted native species in the wild. But good news! Campaigners are encouraging fishing of the aggressive beasts, which go very well in a salad.
- Farming can also make life easy for invaders. In Central America, intensive farming of coffee uses pesticides, which have disrupted the food chain and allowed the spread of rust fungus. A poor coffee harvest this year led Guatemala to declare a state of emergency.
- In 1935, the poisonous cane toad was imported to Australia to eat crop pests, but now it is the pest. There are hundreds of millions of them, roaming the outback with no predators.
- Nowadays, the International Organisation for Biological Control are the people looking for safe species to use in pest control. At the moment, they are looking for a natural enemy of the cabbage moth, but one that won't go on to become a problem itself.
- To prevent the spread of pests, countries have strict customs rules. All planes arriving in Australia, for example, are sprayed with insecticide.
- You can help researchers map the infestation of Spanish slugs by sending your sightings to slugwatch.co.uk.
10:13 - Bacteriophage therapy for C. diff
Bacteriophage therapy for C. diff
with Martha Clokie, Leicester University
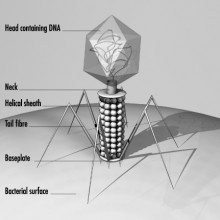
Microbiologist Martha Clokie, from Leicester University explained to Chris Smith how she is developing 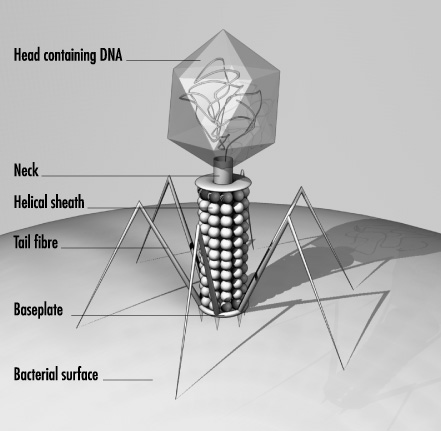 viruses called phages to attack the Clostridium difficile - C. diff - bacterium, which can cause severe diarrhoeal disease, particularly in elderly patients in hospital...
viruses called phages to attack the Clostridium difficile - C. diff - bacterium, which can cause severe diarrhoeal disease, particularly in elderly patients in hospital...
Martha - Clostridium difficile is found naturally in the gut. What happens to patients is that, often, when they're given antibiotics to treat another infection, all the normal bacteria present in the gut are destroyed apart from Clostridium which can then massively expand in the gut area and it releases toxins, or poisons, which then cause very, very severe diarrhoea.
Chris - So, in fact, it's there already, but you effectively clear the way by destroying all the healthy or good bacteria with the antibiotics and that enables it to gain a toehold?
Martha - Yes, that's right. Well, often it can be present. In adults, perhaps about 5% of adults carry it normally. But one of the main problems of Clostridium difficile is that it makes spores and these spores, once they're in a hospital environment, are very, very difficult to remove. So, that's why outbreaks are often associated with hospitals.
Chris - So, you have a symptomatic patient, who puts some spores into the hospital environment and then another person comes along - even if they weren't carrying C. diff to start with - picks up the spores and then the toxic combo of those spores plus a whole host of antibiotics for another infection wipes out their normal bugs and enables them to then get colonized; and then get a major problem with their C. diff?
Martha - Yes, that is actually what happens.
Chris - So, if someone does get C. diff at the moment, how do we treat it?
Martha - Well, at the moment, there are two antibiotics that are routinely used to treat it - metronidazole and vancomycin. These antibiotics work in most patients, but some patients - they can be okay once they're on these antibiotics, but as soon as you remove the antibiotics, they just relapse and become sick again.
Chris - So, it's almost like the balance of microbes in them isn't quite right so they keep getting the C. diff coming back again?
Martha - Yeah, that's right.
Chris - So, what are you doing to try to tackle this?
Martha - What we're doing is trying to find a new way to treat it. What we're looking for are viruses that naturally infect Clostridium difficile. Clostridium is a bacterium, so it's a single-celled organism, and viruses are much, much smaller, and they only exist when they're infecting another host. In the same way, you or I can have flu, Clostridium difficile has viruses that will only infect it. So, what we've been doing is actually trying to find these viruses in the environment and then see if they're effective to treat the patients who had Clostridium difficile.
Chris - This is called 'phage therapy', isn't it? This is not a new idea. Because, looking back in history before we had good quality antibiotics, people were trying to do this for a whole host of infections, particularly in Russia, weren't they?
Martha - That's right. Viruses that infect bacteria were first of all found nearly 100 years ago, and in the '20s, '30s and '40s, they were developed for many, many different diseases. Actually, in places like Georgia, they still use phages to treat many bacterial infections.
Chris - They don't pose any threat to the human that you give them to then?
Martha - No. They're very, very specific. There's no way that they could infect the human cell. They're very specific generally even to the bacterial group. This specificity might be really useful in something like Clostridium difficile, because whereas an antibiotic will suppress Clostridium, it will also kill many other species which you actually don't want to happen, whereas if you take a virus for Clostridium difficile, it would potentially only remove that infection and leave your other bacteria intact.
Chris - I suppose one other major bonus is that when the virus infects the C. diff bacterium, it then makes hundreds, if not, thousands of new bacteriophages programmed to attack C. diff. It will keep going on, amplifying the dose if you like, until it runs out of C. diff to infect and then it will just go away.
Martha - Yes. Well certainly, unlike a conventional antibiotic, when a bacterium is infected by a virus, it will raise perhaps 100 new viruses and they will then as you say, sort of search through the rest of the infected area until they run out of hosts to kill.
Chris - So, have you got some bacteriophages that will attack C. diff in this way?
Martha - Yes, we have, after fairly extensive searching, and a good team of PhD students and post docs. We eventually have come up with a very promising set of viruses that infect Clostridium difficile.
Chris - So, how will you deploy them? What's the next step?
Martha - Well at the moment, having got the viruses, we've shown that they work individually. So, we're now spending about a year or so, trying to design the optimal combination of viruses. Once we've done that, what we hope to be able to do is to then put them into a capsule which would then be able to pass through the stomach and then when it gets to the colon, the capsule would dissolve and the viruses would be released in the targeted way.
Chris - What about the fact that you mentioned that these bacteriophages - these viruses - are extremely selective and specific for the bugs they will infect. What about the fact that there may be different strains of C. diff out there in different people? Will you end up with effectively lots of C. diff that is immune to your phages?
Martha - Well, what we're doing at the moment and we're still in this work - I should emphasise we're still at quite a preliminary stage; we have the viruses and we're now testing which strains they infect. But what we intend to do is find the smallest number of viruses that infect the most number of strains that are circulating in patients. So, at any one time, there are a subset of strains that are causing the most number of cases of disease. So, what we'd hope to find is a little, small set of viruses that can infect these strains. But one of the real attractions of phage therapy is that as new strains come into circulation, we'll be able to then just find other viruses that are then effective in these new strains. So, you don't have to go right back to the drawing board. We just have to go back into our bank of viruses and find one that's effective on these strains...
16:38 - Flu attacks immune memory cells
Flu attacks immune memory cells
Scientists at the Whitehead Institute in the US have made a step forward in understanding why the flu virus is so deadly. And they've done it in a very 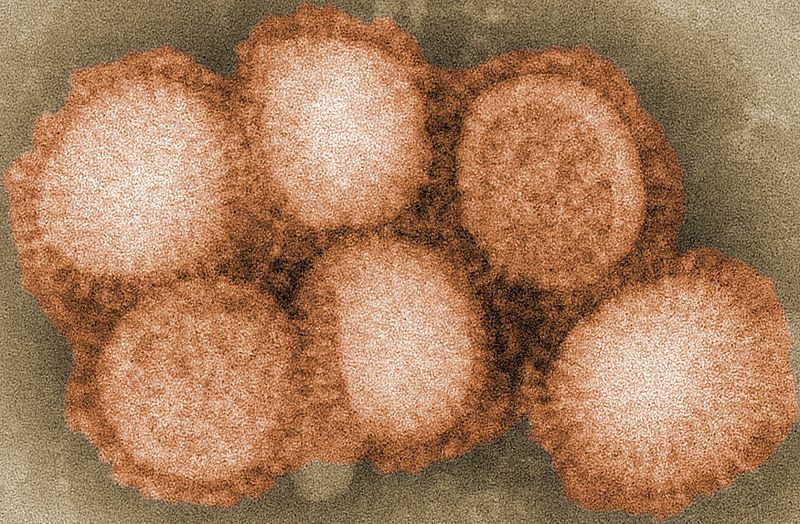 clever way.
clever way.
When we're infected by the influenza virus, our immune system generates cells, known as B cells, that produce antibodies - special proteins that recognise the virus and neutralise it. A subset of these cells take up residence in the lungs, to help fight off the chances of reinfection by the virus through inhaling it.
These cells, known as memory B cells, are particularly good at recognising the virus, so they can spring into action and make more antibodies at the first sign of infection. But the scientists discovered that the flu virus infects and kills these cells in the lungs, making it harder to fight off the infection.
Unfortunately, it's very difficult to purify flu-infected B cells, so they had to take a more roundabout approach, using cloning techniques. They took the DNA - the nucleus - from a mouse B cell that could specifically recognise the flu virus, and put it into a mouse egg cell that had had its own DNA removed. This then created a mouse in which all the B cells can only recognise flu, so they could easily study the infection.
They discovered that the virus infected the flu-specific memory B cells in the lungs and killed them, while flu-specific B cells in another part of the body - the lymph nodes - weren't destroyed. So it looks like the flu virus specifically targets and kills the memory cells that are aiming to protect the body against reinfection from inhaled viruses, giving it another chance to gain a foothold before the rest of the immune system kicks in to tackle it.
The new results, published in Nature, help to explain why flu is so infectious, and could help with the design of vaccines, both for the annual flu shot or entirely new types of preventive approaches. And the techniques the lab have developed could be applied to studying how other viruses interact with cells in the immune system, which could bring health benefits in other diseases.

19:53 - What does sleep do?
What does sleep do?
The fundamental role of sleep might be to flush the brain of more than just unwanted thoughts, scientists have concluded.
Despite decades of effort, and clear evidence  that a lack of sleep is fatal for flies, mice or humans, the role of the process that dominates at least a third of our lives has remained unclear.
that a lack of sleep is fatal for flies, mice or humans, the role of the process that dominates at least a third of our lives has remained unclear.
Now University of Rochester researcher Maiken Nedergard and her colleagues, writing in Science, have found evidence that at least part of the role of sleep might be to cleanse the brain of a toxic build-up of metabolic rubbish that accumulates during the day.
Working with mice, the team injected dyes into the cerebrospinal fluid that bathes the brain and compared where the dye went when the animal was asleep versus when it was awake.
In sleeping animals, they found, the dye rapidly infiltrated throughout the brain tissue, indicating a rapid flux of the cerebrospinal fluid around the brain cells.
But in animals that were awake when the dye was administered, the spread was greatly reduced. The same result was achieved when sleep was induced with anaesthetic agents.
Next, the team looked at molecules like beta-amyloid, the protein which accumulates pathologically in the brains of patients with Alzheimer's Disease.
By continuously collecting small samples of the cerebrospinal fluid surrounding brain cells, they found beta-amyloid proteins were cleared twice as fast from the brains of animals when they slept.
The effect appears to be driven by activity in nerve cells that produce the nerve transmitter chemical noradrenaline and signal sleep or awakes states. This appears to open up the flow of cerebrospinal fluid by allowing the volume of the space around brain cells to increase by up to 60%.
Thus, the team say, "the restorative function of sleep may be due to the switching of the brain into a functional state that facilitates the clearance of degradation products of neural activity that accumulate during wakefulness..."
22:36 - Antibiotic resistance in hospitals
Antibiotic resistance in hospitals
with Nick Brown, Addenbrooke's Hospital, Cambridge
In the first half of 2013, the UK's Chief Medical Officer, Dame Sally Davies described antibiotic resistant bacteria as a "ticking time bomb" which could make routine operations deadly in just 20 years. Kat Arney spoke with Dr Nick Brown, consultant medical microbiologist and lead infection control doctor at Addenbrooke's Hospital in Cambridge...
Nick - Well of course, antibiotic resistance is an entirely natural phenomenon and something that we could consider to be entirely predictable because bacteria had been around for tens of thousands of years. Most of the antibiotics that we have today are products that were developed from naturally occurring substances in the environment. So, bacteria have developed ways of overcoming their activity. But if an organism is able to become resistant to an antibiotic then it won't respond to treatment with that agent. Therefore, the outcome from infections is more severe. People might not be able to go home from hospital for example. They might stay in the hospital longer. The results of the treatment might not be as effective and some things that we might consider to be common as part of everyday modern healthcare, for example, total hip replacement or cancer treatments might become impossible if we have no antibiotics to treat the infections that might complicate them.
Kat - Now is this a particularly recent problem or has it been going on for some time?
Nick - Antibiotic resistance was described very, very soon after the first antibiotics were developed. But in fact, bacteria have had the ability to become resistant for far longer than that. So, there have been studies in Canada on permafrost for example and studies on soil that was taken from around the roots of plants that were harvested in the 17th century that showed that the bacteria in these samples had genes that had a potential to confer resistance to antibiotics that are used today. So certainly, the organisms have the ability to develop resistance very quickly.
Kat - Can you paint me a bit of a picture of the actual scale of the problem?
Nick - Well, it's very difficult to give just a single absolute number of course because the problem varies from organism to another and from one place to another, not only in the community and in the hospital, but also, from one country to another. But if we take some specific examples for common infections that cause for example, cystitis or urinary tract infection, now, there are organisms that are resistant to all of the usual oral antibiotics that GPs in this country have available to them. People are having to be admitted to hospital to have intravenous treatment. In other parts of the world, common infections such as pneumonia or other chest infections can't be treated with the penicillin group of antibiotics because the most common organism that causes this infection, Streptococcus pneumoniae is becoming resistant. So, it varies from one place to another, one organism to another. But what is common I think across the whole patch is that resistance is becoming more common. Not only that, of course, the other big issue is that we don't have any new antibiotics that are coming along to take the place of the ones that are becoming less effective.
Kat - We'll be covering later on in the show about some of the new approaches that people are taking, but I just wanted to explore maybe why we're seeing such a problem now. Because you hear in the papers that it's because doctors have been giving out antibiotics like sweeties, how much has overuse of antibiotics led to this resistance challenge that we have now?
Nick - I think that it is certainly an issue. If we think of the main strategies that we have for controlling resistance then clearly, making the use of antibiotics more appropriate is one key area to - if you like, protect what we've got. But overuse of antibiotics is something that occurs very widely and perhaps also, in some respects, more important is the variation in antibiotic use, for example between one GP and another and between one country and another, and that's not easily explainable.
Kat - So, is this the kind of approach that we need to take across borders? Because I guess people travel so much, that we're just spreading infections all around the world?
Nick - Absolutely, this is key. In some areas of the world, the problem is being taken much more seriously than in others. That is one of the key issues that organisations such as WHO are taking very seriously at the moment.
Kat - If we don't take it seriously, if we don't properly get a lid on bacterial resistance, what are some of the costs that we could face? You talked about making surgery very difficult, but in terms of the financial costs and human costs.
Nick - Absolutely in both and we forget, I think how much we've become to rely on antibiotics in all aspects of healthcare. So, not only the traditional infections if you like - the chest infections, the skin infections, urine infections, but the treatment of cancers where survival has improved dramatically. But the aggressive chemotherapy we now give people make them much more vulnerable to infection. Bone marrow transplantation, organ transplantation, liver, kidney, heart, lung transplants for example would not be possible if you didn't have any antibiotics to treat the infections that occur after them.
Kat - And we're going to talk more about designing specific strategies, but broadly, what would you like to see happen very, very quickly to help get on top of this problem?
Nick - Well, in the introduction, we've already discussed the Chief Medical Officer's Annual Report and in that, to summarise, the two key areas of activity really, the first - to protect what we've got, to make better use of antibiotics to prevent the emergence of new resistance, and to prevent the resistance that is already occurring spreading more widely. And then the second area is to try and regenerate the pipeline for drug discovery of new antibiotics because although there was a golden era of antibiotic development until about 1968 when 15 classes of antibiotic were discovered and developed. Since then there have only been 5 classes of antibiotic discovered and in fact, you could argue that some of those were actually just modifications of ones that had been there before.
29:43 - Sequencing the Microbiome
Sequencing the Microbiome
with Jack Gilbert, Argonne National Laboratory, Chicago
New technologies that can rapidly read the DNA sequences of bacteria can be used to study the way individual bugs move about, how they affect our health and how we can minimise the risks of them becoming resistant to antibiotics. Chris Smith spoke with Jack Gilbert, from Argonne National  Laboratory in Chicago to discover how...
Laboratory in Chicago to discover how...
Jack - You're shedding about 15 million bacterial cells into the immediate environment around you every minute. Those 15 million cells comprise mostly of bacteria associated with skin, but they also come from your nose and your mouth, and interestingly, even from your gastrointestinal system via your trousers and on to these seats you're sitting on. So, you leave a signature wherever you go. And those bacteria land on the surface, most of them die, but some of them go dormant or they proliferate, then they can be transmitted from you to another person.
Chris - Which means if we were in a hospital and another person comes into that part of the hospital where someone with a certain type of bacteria has been, there is, therefore, the prospect for them to pick up those bugs.
Jack - Precisely and we believe this is the main element of transmission. There's a hypothesis which stipulates that it's really just people touching each other. So, when you go and shake hands or you hug, or the doctor comes and talks to you and pats you on the shoulder, that's where that transmission comes from. There's an element which believes it is just in the air - i.e. the bacteria living on little bio-aerosols, these little particles in the air and you inhale them and that's where you get the infection from. And there's groups that believe that the bacteria live on surfaces. You go along and touch that surface after someone else has touched it and you're going to catch that bacterium. But there are literally tens of thousands or hundreds of thousands of bacterial species living alongside the bad ones that make you sick. We don't understand what role they play in promoting or suppressing the transmission of those pathogens.
Chris - So, there are good guys, there are bad guys. The good guys might stop you catching the bad guys, then they also do other things to your body to keep you healthy. What don't we know fundamentally and how can we find out the answers to those questions?
Jack - Only in the last, say, 10 years have we had technology which enables us to rapidly and adequately describe these environments and these communities. So, we're just touching the surface. In a human body, there are 100 billion bacterial cells and we have spent the last 5 years cataloguing those extensively. But without much benefits, we're still making literally making a catalogue, a shopping list of the bacteria that live within us and what they do. We are finding extraordinary things. They influence our behaviour. They influence our mood. They can determine whether you are depressed or anxious. They also influence our disease state, i.e. if you have a healthy bacterial community in your intestine, in your mouth then that can prevent you from getting a pathogenic infection. They have to increase or decrease your metabolic clock inside your body. So jetlag, we can potentially cure by putting the right types of bacteria back inside your gut.
Chris - How can we use this understanding to make sure we either don't get antimicrobial resistance or to mitigate what we already have got?
Jack - That's a very complicated question and that's exactly what we're here today to try and uncover. How do we design the best experiments possible so we can determine exactly why certain bacterial communities may affect that process. One of the fundamental problems that we have is that we don't understand which bacteria are out there and how they interact together. So, by defining those links, saying how bacteria talk to each other daily, finding out how they communicate and how they respond to that communication could change the way we think about disease infection. Especially antimicrobial resistance because it's when those bacteria talk to each other that they transmit that resistance between each other. So, we need to understand that fundamentally. We believe that the hospital environment "may" be - I stress "may" - may be a reservoir of antimicrobial resistance.
Chris - So, how can we answer those sorts of questions? What are we armed with in terms of the tools to do this research now that we didn't have in the last 10 years?
Jack - We've fundamentally increased our ability to sequence the genomes of these organisms. Much the same as the Human Genome Project, we can sequence a bacterial genome for $300 now and that data stream is helping us to ask - where before, we would've analysed 20 or 30 patients, now, we can analyse 20 or 30,000 patients. That ability changes our capacity to do statistical analysis. As any scientist will tell you, if the statistics don't hold up, it's not true. So, we need that power. We need that number of observations to really get at the statistical integrity.
Chris - So, you can use DNA technology to probe what are these bacteria which I presume is going to be good news, because not all bacteria grow where previously we tried to grow things in dishes. We would've missed some, so that may be a factor too.
Jack - So, this is exactly the idea that some organisms grow really well in the lab and that's what we've been looking at. But there are circumstantial evidence which suggests that a lot of the diseases we encounter, we don't know what bacteria or what virus causes that disease. So, these kinds of analyses that look at bacteria in situ with sequencing technologies and we can identify bacteria which are always present when that person has that illness, always present when multiple people have that illness. It helps us to identify new diseases.
Chris - Can it also help us to identify who may be at risk of a disease?
Jack - The concept of this is, that the bacterial community in your body undergo what we would call dysbiosis. They're no longer in symbiotic union with us. When that symbiotic relationship breaks down, we are open to all kinds of mischief. Different pathogens can infect us. Our metabolic states in our guts can change. Our liver condition can change. That has serious implications for our health and well-being in a hospital.
Chris - Jack Gilbert from Argonne National Laboratory in Chicago and thank you also to the foreign office's, Jack Westwood. He is based at Science and Innovation Network in Chicago. He invited us to that meeting.
36:13 - Preventing antibiotic resistance
Preventing antibiotic resistance
with John Wain, University of East Anglia
Rapid DNA-sequencing technology can be used to identify quickly the bacteria responsible for causing an infection, meaning that narrow-spectrum targeted drug treatments, with inevitably fewer side effects, can be administered to patients, as John Wain, Professor of Microbiology at the University of East Anglia, explains to Kat Arney...
Kat - How are bacterial infections currently identified and how long does that take?
John - So, the identification currently is through culture at the microbiology lab. So, we actually have to grow them. Some people call it chemical gardening, but it's a matter of days before we can actually get a result. What that means is that when you go to the GP, the doctor has given you antibiotics on clinical grounds. So, they take a look at you, decide if you've got an infection or not and then the decision to actually give antibiotics is taken and then the samples are taken and the diagnostic results come back.
Kat - So, it's a bit of a shooting in the dark approach and if you get the wrong one to start with, it could be really serious.
John - That's why this idea of broad spectrum antibiotics tends to be quite popular. So obviously, if you're - as you put it - shooting in the dark then you want something very broad so that you make sure that you kill what it is that you are actually shooting at.
Kat - So, how are we using new genetic techniques to actually try and get down to a precision rifle at these bugs?
John - Okay, so obviously there are problems with using broad spectrum. There's the problem of collateral damage as we heard with the Clostridium difficile - if you interfere with the gut flora, you're likely to get overgrowth of a pathogen. So, this specific idea is that we just get the pathogen and we don't hit the normal flora. So, there are two ways that we can overcome that. The first is, with good diagnostics and that's where these new technologies in sequencing can really help. So, we've heard about being able to recognise the microbiome. You can also recognise the pathogens that are causing infection if you take the right samples, and if you can get that sequencing out quickly enough then you can get a real-time evidence in so that you can actually use targeted antibiotics.
Kat - And how quickly are we talking about and how quickly could we do that nowadays and how quickly do you think it will become possible in the future?
John - The quickest we could do it at the moment is we can do two large scale sequencing runs a day. 24 to 48 hours realistically is the quickest you can do it at the moment. But that's on a large machine which is laboratory based. There are new technologies coming out which look sort of USB sticks that you put into a computer. Those will, I believe in the not too distant future, be giving us results much more quickly.
Kat - So, that's something that's really positive. But then what can we do with this information that can actually make treatment more effective and particularly overcome this problem of resistance?
John - So, the other problem to using targeted antibiotics is developing targeted antibiotics. So, there are lots of chemicals out there, lots of small molecules, lot of compounds that hit bacteria. So, we know we can kill this bacteria, but the problem is, taking those hits to what the chemist call hit optimisation. So, choosing which of those chemical compounds to actually take forward. Then using high throughput sequencing technologies, we can now X-ray a bacterium, we can look right inside that bacterium and actually work out exactly what these small molecules are doing. That allows the chemists to optimise them more effectively so that we can choose which ones to put into lead op.
Kat - I guess it's a lot similar way to the kind of targeted therapies in cancer that we're seeing nowadays isn't it?
John - Yes, it is very much so. It's also obviously has similarities with what we're hearing about with the bacteriophage therapies.
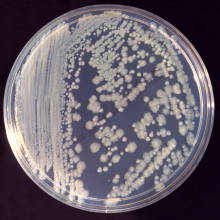
40:15 - Discovering new antibiotics
Discovering new antibiotics
with David Williams, CEO, Discuva
New antibiotics, which can tackle specific bacterial infections, are urgently needed to overcome rising rates of resistance. But how are these  new drugs developed? One Cambridge-based company, Discuva, are using genetic techniques to identify new drug compounds to which bacteria struggle to develop resistance. David Williams, CEO of Discuva, took Ginny Smith to his lab to demonstrate how they go about finding potential new drugs...
new drugs developed? One Cambridge-based company, Discuva, are using genetic techniques to identify new drug compounds to which bacteria struggle to develop resistance. David Williams, CEO of Discuva, took Ginny Smith to his lab to demonstrate how they go about finding potential new drugs...
Ginny - So, you're looking for a compound that might work as a drug to kill bacteria. Where do you even start? There must be so many compounds out there.
David - There's about 10 million compounds you can acquire. So, what we do is we look at those computationally to look for chemical qualities that we think will allow them to penetrate a bacterial cell. We screen them against different bacteria, that are pathogens in our hospital today.
Ginny - And can you show me how you do that screening?
David - Yeah, sure. We have here a microtitre plate and this is essentially loads of test tubes joined together and we put our compounds, our chemicals, in each of these individual test tubes, and we put bacteria on top of them, and we grow them over time. The bacteria that aren't affected by the compound grow and become cloudy in solution and for the ones that are prevented from growing, there's a clear solution.
Ginny - How many wells are there in each of those?
David - In each plate, there's 386 wells.
Ginny - Wow! So you can look at 386 different possible drugs at the same time.
David - That's correct.
Ginny - Can you show me the computer system that actually tells you which ones have worked?
David - Yeah, I can take you over there now.
Ginny - Okay, so it's a big grey cupboard basically.
David - We have a reader inside here - a plate reader - which will scan through 50 plates at once.
Ginny - And then there's a computer next to it. It's got lots of little circles on it and they're all different colours. What does that mean?
David - Each of the coloured spots represents an individual well in the plate.
Ginny - Okay, so there's a load of blue spots down one side. What do they mean?
David - Well, that's where we don't have any bacteria in the well at all. They just have the growth medium in them. So, the blue means there's been no bacterial growth, so no cloudiness of the solution.
Ginny - So, what you're looking for is one of the other spots that has bacteria in it, but has a different medium to be blue as well because that would mean the bacteria hadn't grown.
David - That's correct. At the moment, they're green in colour which means there's a lot of growth.
Ginny - So, none of these would be possible candidates for a drug.
David - This particular plate, no, not at the moment. This is a bacterium called Pseudomonas. This particular bacteria is a really difficult pathogen to treat. It causes various blood infections and respiratory infections.
Ginny - Although there aren't any here that might be candidates, you do often get them coming up. How do you figure out if this is going to be a really effective antibacterial treatment or if it's just going to work only in some circumstances and not for very long?
David - Well, we've got a very neat technology that we've developed. We had to take some of these pathogens that are problems and engineer them. We've engineered them in such a way that we can use them as mini computers to tell us what's happening to a compound when they're treated with it.
A bacterium has a piece of DNA, its chromosome, which is 8 million base pairs long. We put individual tags in different bacteria at every position in that genome and those tags are pieces of DNA that can disrupt genes or activate genes in the position that the tag has been placed.
We incubate all of those bacteria with our compounds and without our compound. After we've grown those bacteria, we sequence the bacteria that've gone into the experiment and the bacteria that've come out of the experiment. It's the difference between the mutants that we see that gives us the information on what the compounds are doing.
So, it can tell us exactly what component inside the cell the compound is binding to and affecting, and it can tell us what all the potential resistance mechanisms could be for that compound. That's really important because we don't want to develop new antibiotics that are going to develop resistance quickly in the clinic.
We want to be working on compounds that can last a long time once they're on the market and in patients.
Ginny - So, you're essentially trying to speed up what would happen in nature once people started being treated with this new compound because bacteria are very fast at evolving and can develop resistance quite quickly. So, is that what your technique is doing, sort of trying to speed that up and see what would happen in real life?
David - Yes, absolutely right. We can do what nature does in 10 years in a week.
Ginny - Brilliant! So then you can pick out the ones that it's hardest for the bacteria to evolve resistance towards and focus on those for the future.
David - That's exactly what we do and all of our compounds that we're working on for new drugs have those qualities in built.
Ginny - You're talking about 8 million base pairs, 8 million different types of bacteria and then you're sequencing the genome for each of those. There must be huge amounts of data. Does this take days and days to do and to process?
David - Three or four years ago, it was impossible to do this. It's really the rapid advances in next generation sequencing technology that have enabled us to be able to do this. This particular instrument can give us sequence information back from 200 million different sequencing reactions in just a few hours. The information that we generate is huge. I mean, we generate in a week, more information than has been generated in the whole London Stock Exchange in its history.
Ginny - Wow! Have any of your drugs reached clinical trials yet?
David - No, we're relatively new company. We've only been in the laboratory for just under 2 years. For us to pursue any of our drugs to clinic, it is going to be a minimum time of 3 years...

46:13 - Resensitising bacteria to old agents
Resensitising bacteria to old agents
with Mike Davies, Blueberry Therapeutics
 With rising rates of antimicrobial resistance, one company, Blueberry Therapeutics, has been working on a way to give a new lease of life to old drugs, by re-sensitising them to the antibiotics we already do have, as Blueberry Co-founder, and former surgeon, Mike Davies explains to Chris Smith...
With rising rates of antimicrobial resistance, one company, Blueberry Therapeutics, has been working on a way to give a new lease of life to old drugs, by re-sensitising them to the antibiotics we already do have, as Blueberry Co-founder, and former surgeon, Mike Davies explains to Chris Smith...
Chris - So, you've decided to swap your scalpel for a sort of molecular scalpel in this instant.
Mike - Indeed, yes. Well, we're Blueberry Therapeutics. What we're doing, is to basically inhibit the resistance of resistant bacteria. So, we effectively make them sensitive to current antibiotics, the ones that they have become resistant to. We do that with a very clever technique using a nanoparticle delivery. We have a nanopolymer that basically wraps itself around certain things, certain drugs, certain proteins and things, and delivers them into the bacterial cells. So, we have developed what are called aptamers and then specifically this area, something called an affimer which is really a tiny, tiny little antibody, really incredibly small, related to an antibody. These are taken up by the nanopolymer into what's called a nanoparticle and effectively delivered into the bacterial cell. We're targeting the way that those bacteria actually inhibit antibiotics. There are over 200 ways that bacteria inhibit antibiotics and stop them from working. We're literally switching those off.
Chris - So, what happens then if the bacteria mutate because one of the things that bacteria do is they change all the time. So, why can't they just change to get around what you're throwing at them?
Mike - Well, that's a very good question and of course, we will have to look for that as we develop these compounds, but first of all, they haven't done that before because there haven't been these ways of blocking really effectively the resistance mechanisms. We can probably keep pace with that because we have these small proteins, that we call affimers, and they are literally a small number of amino acids - the things that build up proteins long. And they're built like a tiny antibody as I say. So, they've got two arms that we can change by one amino acid and it will literally change how it binds. Now, with that process, the thought is, and it's a big picture for the future, but the thought is, as a bacterium or bacteria change and become resistant perhaps to one of our affimers, we can rapidly change it so we develop a new way of blocking the new resistance.
Chris - So, if you're putting in something that is a protein though which is what your treatment is, is there not a danger that a person's immune system will react to that and then you'll get an immune response? You'll make your own antibodies to your drug.
Mike - Well, that's again a very, very good question and something that we would carefully look for in the development. Although we we will start off by the topical delivery of these aptamers, so we'll start off in MRSA infected wounds, such as venous leg ulcers and diabetic foot ulcers. And the affimers that we give are targeted directly at the bacteria. We doubt if we will see an immune response. We will have to look for that and it's a very important part of the drug development.

Can dogs catch and transmit human viruses?
- Previous Science of Baking
- Next Can we make a fake brain?










Comments
Add a comment